1D9O
 
 | |
1CQ3
 
 | | STRUCTURE OF A SOLUBLE SECRETED CHEMOKINE INHIBITOR, VCCI, FROM COWPOX VIRUS | | Descriptor: | VIRAL CHEMOKINE INHIBITOR | | Authors: | Carfi, A, Smith, C.A, Smolak, P.J, McGrew, J, Wiley, D.C. | | Deposit date: | 1999-08-05 | | Release date: | 1999-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D9M
 
 | |
1QMP
 
 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
3PVA
 
 | | PENICILLIN V ACYLASE FROM B. SPHAERICUS | | Descriptor: | PROTEIN (PENICILLIN V ACYLASE) | | Authors: | Suresh, C.G, Pundle, A.V, Rao, K.N, Sivaraman, H, Brannigan, J.A, Mcvey, C.E, Verma, C.S, Dauter, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members.
Nat.Struct.Biol., 6, 1999
|
|
1CQT
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX CONTAINING AN OCA-B PEPTIDE, THE OCT-1 POU DOMAIN, AND AN OCTAMER ELEMENT | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), POU DOMAIN, ... | | Authors: | Chasman, D.I, Cepek, K, Sharp, P.A, Pabo, C.O. | | Deposit date: | 1999-08-11 | | Release date: | 1999-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface.
Genes Dev., 13, 1999
|
|
1D3U
 
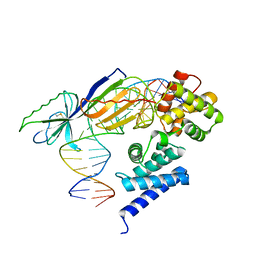 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/BRE+TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA 23-MER: BRE+TATA-BOX, DNA 24-MER: BRE+TATA-BOX, TATA-BINDING PROTEIN, ... | | Authors: | Littlefield, O, Korkhin, Y, Sigler, P.B. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the oriented assembly of a TBP/TFB/promoter complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CTQ
 
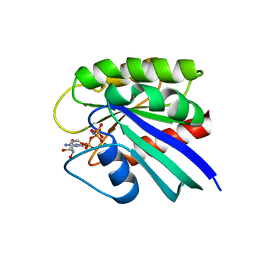 | | STRUCTURE OF P21RAS IN COMPLEX WITH GPPNHP AT 100 K | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (TRANSFORMING PROTEIN P21/H-RAS-1) | | Authors: | Scheidig, A, Burmester, C, Goody, R.S. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins.
Structure Fold.Des., 7, 1999
|
|
1D9V
 
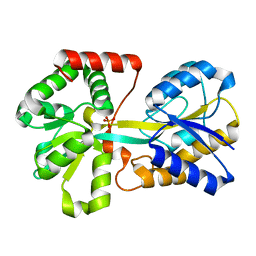 | |
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1EX1
 
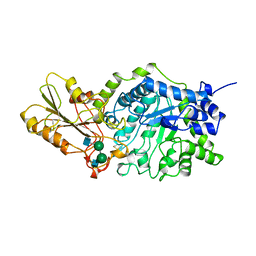 | | BETA-D-GLUCAN EXOHYDROLASE FROM BARLEY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-D-GLUCAN EXOHYDROLASE ISOENZYME EXO1), ... | | Authors: | Varghese, J.N, Hrmova, M, Fincher, G.B. | | Deposit date: | 1998-11-10 | | Release date: | 1999-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase.
Structure Fold.Des., 7, 1999
|
|
2TOD
 
 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
1B0I
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE) | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1998-11-10 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level.
Structure, 6, 1998
|
|
1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1D3V
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH 2(S)-AMINO-6-BORONOHEXANOIC ACID, AN L-ARGININE ANALOG | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Cox, J.D, Kim, N.N, Traish, A.M, Christianson, D.W. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Arginase-boronic acid complex highlights a physiological role in erectile function.
Nat.Struct.Biol., 6, 1999
|
|
1B0U
 
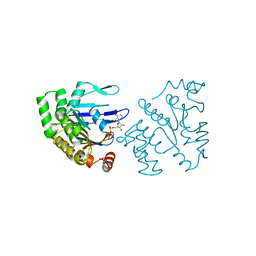 | | ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, HISTIDINE PERMEASE | | Authors: | Hung, L.-W, Wang, I.X, Nikaido, K, Liu, P.-Q, Ames, G.F.-L, Kim, S.-H. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the ATP-binding subunit of an ABC transporter.
Nature, 396, 1998
|
|
1CU6
 
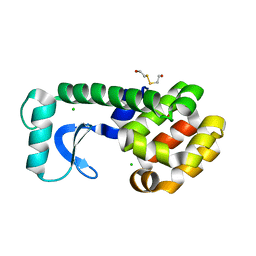 | | T4 LYSOZYME MUTANT L91A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1QS4
 
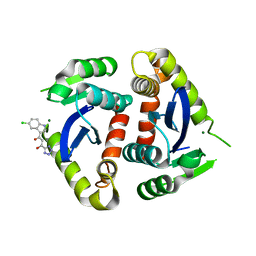 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1SN1
 
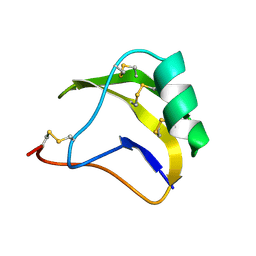 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SN4
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1QU4
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-07-06 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
1DBI
 
 | | CRYSTAL STRUCTURE OF A THERMOSTABLE SERINE PROTEASE | | Descriptor: | AK.1 SERINE PROTEASE, CALCIUM ION, SODIUM ION | | Authors: | Smith, C.A, Toogood, H.S, Baker, H.M, Daniel, R.M, Baker, E.N. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.
J.Mol.Biol., 294, 1999
|
|
1B0L
 
 | | RECOMBINANT HUMAN DIFERRIC LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Baker, E.N, Jameson, G.B, Sun, X. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant human lactoferrin expressed in Aspergillus awamori.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B06
 
 | | SUPEROXIDE DISMUTASE FROM SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Knapp, S, Kardinahl, S, Niklas, H, Tibbelin, G, Schafer, G, Ladenstein, R. | | Deposit date: | 1998-11-16 | | Release date: | 1999-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structure of a superoxide dismutase from the hyperthermophilic archaeon Sulfolobus acidocaldarius at 2.2 A resolution.
J.Mol.Biol., 285, 1999
|
|
