1GQJ
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQK
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQL
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQM
 
 | | The structure of S100A12 in a hexameric form and its proposed role in receptor signalling | | Descriptor: | CALCIUM ION, CALGRANULIN C | | Authors: | Moroz, O.V, Antson, A.A, Dodson, E.G, Burrel, H.J, Grist, S.J, Lloyd, R.M, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E, Bronstein, I.B. | | Deposit date: | 2001-11-26 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of S100A12 in a Hexameric Form and its Proposed Role in Receptor Signalling
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GQN
 
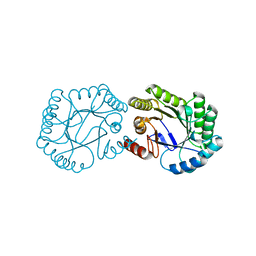 | | Native 3-dehydroquinase from Salmonella typhi | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Lee, W.-H, Perles, L.A, Nagem, R.A.P, Polikarpov, I, Sawyer, L. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparison of Different Crystal Forms of 3-Dehydroquinase from Salmonella Typhi and its Implications for Enzyme Activity
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GQO
 
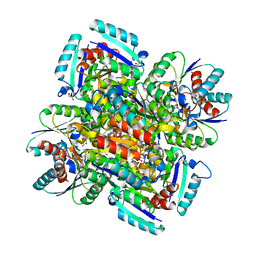 | |
1GQP
 
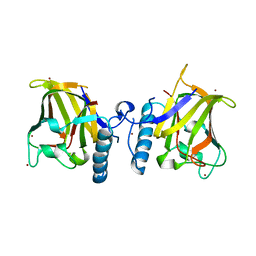 | | APC10/DOC1 SUBUNIT OF S. cerevisiae | | Descriptor: | BROMIDE ION, DOC1/APC10 | | Authors: | Au, S.W.N, Leng, X, Harper, J.W.A.D.E, Barford, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for the Ubiquitination Reaction of the Anaphase-Promoting Complex from the Crystal Structure of the Doc1/Apc10 Subunit.
J.Mol.Biol., 316, 2002
|
|
1GQQ
 
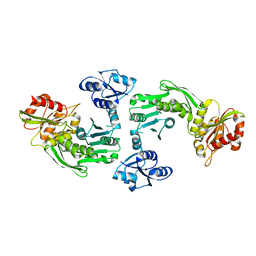 | |
1GQR
 
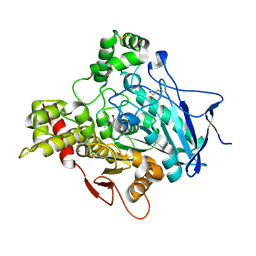 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH RIVASTIGMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE, ... | | Authors: | Raves, M.L, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1GQS
 
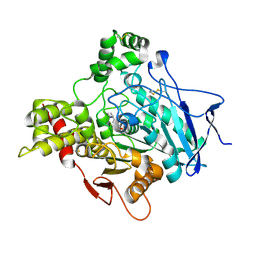 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH NAP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE | | Authors: | Bar-on, P, Millard, C.B, Harel, M, Dvir, H, Enz, A, Sussman, J.L, Silman, I. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1GQT
 
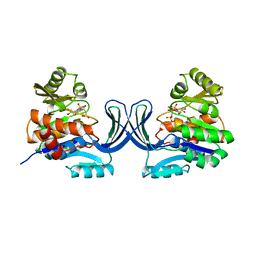 | |
1GQU
 
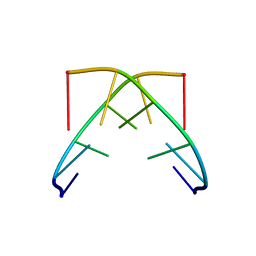 | |
1GQV
 
 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
1GQW
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Ryle, M.J, Clifton, I.J, Dunning-Hotopp, J.C, Lloyd, J.S, Burzlaff, N.I, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates
Biochemistry, 41, 2002
|
|
1GQY
 
 | | MURC - CRYSTAL STRUCTURE OF THE ENZYME FROM HAEMOPHILUS INFLUENZAE COMPLEXED WITH AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE | | Authors: | Skarzynski, T, Cleasby, A, Domenici, E, Gevi, M, Shaw, J. | | Deposit date: | 2001-12-06 | | Release date: | 2003-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Udp-N-Acetylmuramate-L-Alanine Ligase (Murc) from Haemophilus Influenzae
To be Published
|
|
1GQZ
 
 | |
1GR0
 
 | | myo-inositol 1-phosphate synthase from Mycobacterium tuberculosis in complex with NAD and zinc. | | Descriptor: | CACODYLATE ION, INOSITOL-3-PHOSPHATE SYNTHASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Norman, R.A, Murray-Rust, J, McDonald, N.Q, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Structure, 10, 2002
|
|
1GR1
 
 | | Structure of Ferredoxin-NADP+ Reductase with Glu 139 replaced by Lys (E139K) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Role of Glutamic Acid 139 of Anabena Ferrodoxin-Nadp+ Reductase in the Interaction with Substrates
Eur.J.Biochem., 269, 2002
|
|
1GR2
 
 | | STRUCTURE OF A GLUTAMATE RECEPTOR LIGAND BINDING CORE (GLUR2) COMPLEXED WITH KAINATE | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, PROTEIN (GLUTAMATE RECEPTOR 2) | | Authors: | Armstrong, N, Sun, Y, Chen, G.Q, Gouaux, E. | | Deposit date: | 1998-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a glutamate-receptor ligand-binding core in complex with kainate.
Nature, 395, 1998
|
|
1GR3
 
 | | Structure of the human collagen X NC1 trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, COLLAGEN X, ... | | Authors: | Bogin, O, Kvansakul, M, Rom, E, Singer, J, Yayon, A, Hohenester, E. | | Deposit date: | 2001-12-12 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight Into Schmid Metaphyseal Chondrodysplasia from the Crystal Structure of the Collagen X Nc1 Domain Trimer.
Structure, 10, 2002
|
|
1GR5
 
 | | Solution Structure of apo GroEL by Cryo-Electron microscopy | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1GR7
 
 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
1GRA
 
 | |
1GRB
 
 | |
1GRC
 
 | |
