3UGF
 
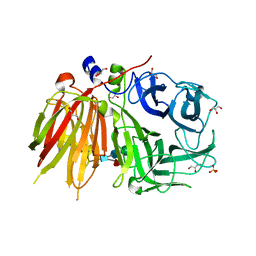 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UC8
 
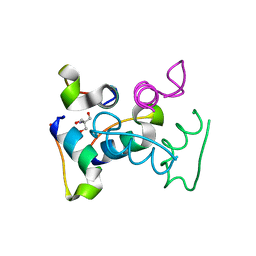 | | Trp-cage cyclo-TC1 - tetragonal crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, cyclo-TC1 | | Authors: | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2BUB
 
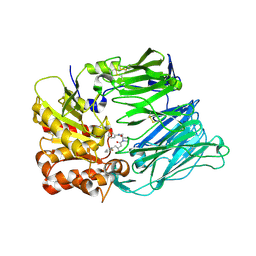 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIPEPTIDYL PEPTIDASE 4, N-({(2S)-1-[(3R)-3-AMINO-4-(2-FLUOROPHENYL)BUTANOYL]PYRROLIDIN-2-YL}METHYL)BENZAMIDE | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | Deposit date: | 2005-06-09 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
3UCS
 
 | |
2C0Q
 
 | | non-aged form of mouse acetylcholinesterase inhibited by tabun | | Descriptor: | ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL, R-ETHYL N,N-DIMETHYLPHOSPHONAMIDATE | | Authors: | Ekstrom, F, Akfur, C, Tunemalm, A.-K, Lundberg, S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Changes of Phenylalanine 338 and Histidine 447 Revealed by the Crystal Structures of Tabun-Inhibited Murine Acetylcholinesterase.
Biochemistry, 45, 2006
|
|
3UIX
 
 | |
3UJE
 
 | | Asymmetric complex of human neuron specific enolase-3-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
7N31
 
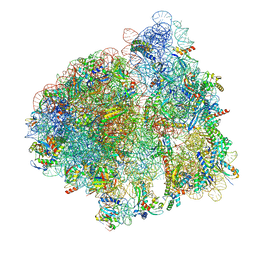 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
6A1N
 
 | | Mandelate oxidase mutant-Y128F with (2R,3S)-3-fluoro-2-hydroxy-3-phenylpropanoic acid | | Descriptor: | (2R,3S)-3-fluoro-2-hydroxy-3-phenylpropanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
1WF7
 
 | | Solution structure of the PDZ domain of Enigma homologue protein | | Descriptor: | Enigma homologue protein | | Authors: | Qin, X, Tochio, N, Kigawa, T, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Enigma homologue protein
To be published
|
|
3UFL
 
 | | Discovery of Pyrrolidine-based b-Secretase Inhibitors: Lead Advancement through Conformational Design for Maintenance of Ligand Binding Efficiency | | Descriptor: | (1R,4'S)-3,4-dihydro-2H-spiro[naphthalene-1,3'-pyrrolidin]-4'-yl[(2S,4R)-2,4-diphenylpiperidin-1-yl]methanone, Beta-secretase 1, GLYCEROL, ... | | Authors: | Allison, T, Munshi, S, Soisson, S.M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of pyrrolidine-based beta-secretase inhibitors: Lead advancement through conformational design for maintenance of ligand binding efficiency.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6A1W
 
 | | Mandelate oxidase with the enoyl FMN epoxide adduct | | Descriptor: | 1-[(1aR,11R)-11-acetyl-8,9-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]oxazireno[3,2-e]pteridin-11-ium-6(2H)-yl]-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase, MAGNESIUM ION | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
3UKH
 
 | |
2C0P
 
 | | Aged form of mouse acetylcholinesterase inhibited by tabun | | Descriptor: | ACETYLCHOLINESTERASE, ETHYL HYDROGEN PHOSPHONATE | | Authors: | Ekstrom, F, Akfur, C, Tunemalm, A.-K, Lundberg, S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Changes of Phenylalanine 338 and Histidine 447 Revealed by the Crystal Structures of Tabun-Inhibited Murine Acetylcholinesterase.
Biochemistry, 45, 2006
|
|
3UGH
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3DIG
 
 | |
3UIP
 
 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UQP
 
 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
126D
 
 | |
3US4
 
 | |
2C68
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDROPYRAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3UT5
 
 | | Tubulin-Colchicine-Ustiloxin: Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UL4
 
 | | Crystal structure of Coh-OlpA(Cthe_3080)-Doc918(Cthe_0918) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome enzyme, dockerin type I, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
5ZZT
 
 | |
3UM9
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
