1A67
 
 | | CHICKEN EGG WHITE CYSTATIN WILDTYPE, NMR, 16 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1A66
 
 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | Descriptor: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | Authors: | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | Deposit date: | 1998-03-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
1A68
 
 | |
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
2SDF
 
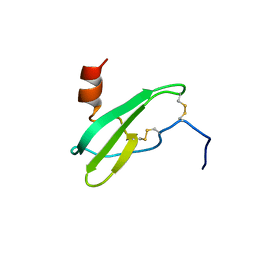 | | SOLUTION NMR STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), 30 STRUCTURES | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-03-07 | | Release date: | 1998-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
3LJR
 
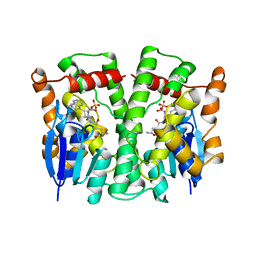 | | GLUTATHIONE TRANSFERASE (THETA CLASS) FROM HUMAN IN COMPLEX WITH THE GLUTATHIONE CONJUGATE OF 1-MENAPHTHYL SULFATE | | Descriptor: | 1-MENAPHTHYL GLUTATHIONE CONJUGATE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
1A69
 
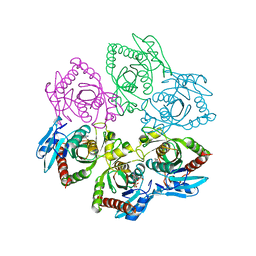 | | PURINE NUCLEOSIDE PHOSPHORYLASE IN COMPLEX WITH FORMYCIN B AND SULPHATE (PHOSPHATE) | | Descriptor: | FORMYCIN B, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Koellner, G, Luic, M, Shugar, D, Saenger, W, Bzowska, A. | | Deposit date: | 1998-03-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ternary complex of E. coli purine nucleoside phosphorylase with formycin B, a structural analogue of the substrate inosine, and phosphate (Sulphate) at 2.1 A resolution.
J.Mol.Biol., 280, 1998
|
|
2LJR
 
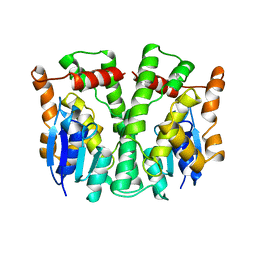 | | GLUTATHIONE TRANSFERASE APO-FORM FROM HUMAN | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
1LJR
 
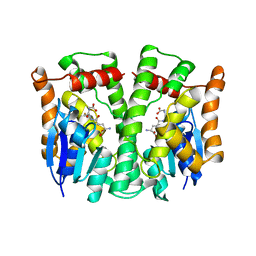 | | GLUTATHIONE TRANSFERASE (HGST T2-2) FROM HUMAN | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
4HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, 25 STRUCTURES | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
1IC1
 
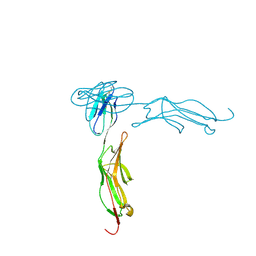 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1NKF
 
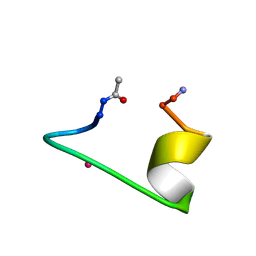 | | CALCIUM-BINDING PEPTIDE, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM-BINDING HEXADECAPEPTIDE, LANTHANUM (III) ION | | Authors: | Sticht, H, Ejchart, A. | | Deposit date: | 1998-03-09 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix nucleation by a calcium-binding peptide loop.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5HCK
 
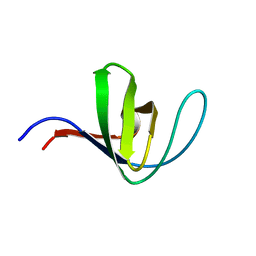 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
1MPS
 
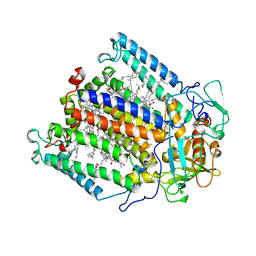 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M 197 REPLACED WITH ARG AND TYR M 177 REPLACED WITH PHE (CHAIN M, Y177F, F197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Mcauley-Hecht, K.E, Fyfe, P.K, Ridge, J.P, Prince, S, Hunter, C.N, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1998-03-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies of wild-type and mutant reaction centers from an antenna-deficient strain of Rhodobacter sphaeroides: monitoring the optical properties of the complex from bacterial cell to crystal.
Biochemistry, 37, 1998
|
|
22GS
 
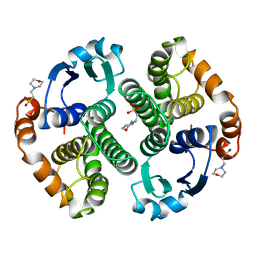 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y49F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A.J. | | Deposit date: | 1998-03-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione.
J.Biol.Chem., 278, 2003
|
|
1UOR
 
 | |
1A7A
 
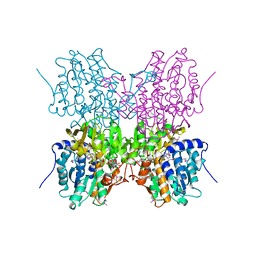 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
1A7B
 
 | |
4LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN NEAT ACETONITRILE, THEN BACK-SOAKED IN WATER | | Descriptor: | LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
3LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN 95% ACETONITRILE-WATER | | Descriptor: | ACETONITRILE, LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
2XAT
 
 | |
1XAT
 
 | |
386D
 
 | | THREE-DIMENSIONAL STRUCTURE AND REACTIVITY OF A PHOTOCHEMICAL CLEAVAGE AGENT BOUND TO DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), N,N-BIS(3-AMINOPROPYL)-2-ANTHRAQUINONESULFONAMIDE | | Authors: | Gasper, S.M, Armitage, B, Shui, X, Hu, G.G, Yu, C, Schuster, G, Williams, L.D. | | Deposit date: | 1998-03-11 | | Release date: | 1998-03-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure and Reactivity of a Photochemical Cleavage Agent Bound to DNA
J.Am.Chem.Soc., 120, 1998
|
|
1TR1
 
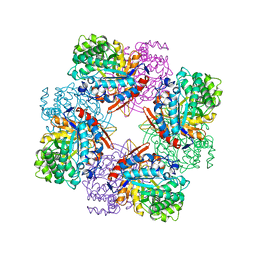 | | CRYSTAL STRUCTURE OF E96K MUTATED BETA-GLUCOSIDASE A FROM BACILLUS POLYMYXA, AN ENZYME WITH INCREASED THERMORESISTANCE | | Descriptor: | BETA-GLUCOSIDASE A, GLYCEROL | | Authors: | Sanz-Aparicio, J, Hermoso, J.A, Martinez-Ripoll, M, Gonzalez-Perez, B, Polaina, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1ICJ
 
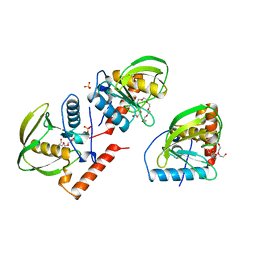 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
