9CTH
 
 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
9EV6
 
 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO), C-terminal truncated construct | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 2024
|
|
8WKY
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9BPF
 
 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BOO
 
 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
8QWV
 
 | |
8QWU
 
 | |
8QWW
 
 | |
9BC6
 
 | | HCN1 M305L with propofol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,6-BIS(1-METHYLETHYL)PHENOL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
9BC7
 
 | | HCN1 M305L holo | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
8UC8
 
 | | HCN1 nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2023-09-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
8UC7
 
 | | HCN1 complex with propofol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,6-BIS(1-METHYLETHYL)PHENOL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2023-09-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
8K7U
 
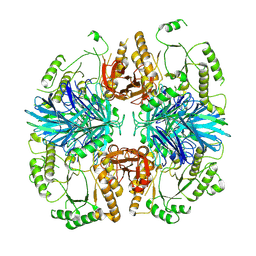 | | the alpha-galactosidase 5 with Cacl2 | | Descriptor: | Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activity-Based Meta proteomics Drives Discovery and Enzymological Characterization of Novel alpha-galactosidases in the Gut Microbiome
To Be Published
|
|
8K7V
 
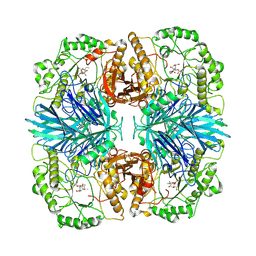 | | the alpha-galactosidase 5 with inhibitor ABP2 | | Descriptor: | 8-azido-1-((1S,2S,3S,4S,5R,6S)-2,3,4-trihydroxy-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptan-7-yl)octan-1-one, Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Activity-Based Meta proteomics Drives Discovery and Enzymological Characterization of Novel alpha-galactosidases in the Gut Microbiome
To Be Published
|
|
9FYP
 
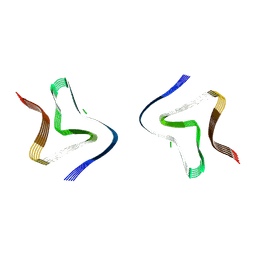 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | Descriptor: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | Authors: | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
8K1A
 
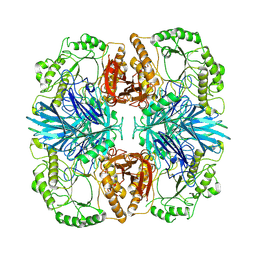 | | the wild-typed alpha-galactosidase 5 | | Descriptor: | Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activity-Based Meta proteomics Drives Discovery and Enzymological Characterization of Novel alpha-galactosidases in the Gut Microbiome
To Be Published
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8RJ6
 
 | | E. coli adenylate kinase in complex with ATP and AMP and Mg2+ as a result of enzymatic AP4A hydrolysis. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
