1F6F
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX BETWEEN OVINE PLACENTAL LACTOGEN AND THE EXTRACELLULAR DOMAIN OF THE RAT PROLACTIN RECEPTOR | | Descriptor: | PLACENTAL LACTOGEN, PROLACTIN RECEPTOR | | Authors: | Elkins, P.A, Christinger, H.W, Sandowski, Y, Sakal, E, Gertler, A, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor.
Nat.Struct.Biol., 7, 2000
|
|
6NX4
 
 | | Structure of the C-terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase (eEF-2K) | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Giles, D.H, Hajredini, F, Dalby, K.N, Ghose, R. | | Deposit date: | 2019-02-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxy-Terminal Tandem Repeat Domain of Eukaryotic Elongation Factor 2 Kinase and Its Role in Substrate Recognition.
J.Mol.Biol., 431, 2019
|
|
2EIF
 
 | | Eukaryotic translation initiation factor 5A from Methanococcus jannaschii | | Descriptor: | PROTEIN (EUKARYOTIC TRANSLATION INITIATION FACTOR 5A) | | Authors: | Kim, K.K, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2MC9
 
 | | Cat r 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Mueller, G, London, R, Ghosh, D. | | Deposit date: | 2013-08-16 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1.
J.Biol.Chem., 289, 2014
|
|
1QAY
 
 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH MEVALONATE AND NAD+ | | Descriptor: | (R)-MEVALONATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3DTV
 
 | | Crystal structure of arylmalonate decarboxylase | | Descriptor: | Arylmalonate decarboxylase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Nakasako, M, Obata, R, Miyamaoto, K, Ohta, H. | | Deposit date: | 2008-07-16 | | Release date: | 2009-07-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inverting the Enantioselectivity of Arylmalonate Decarboxylase Revealed by the Structural Analysis of the Gly74Cys/Cys188Ser Mutant in the Liganded Form
Biochemistry, 49, 2010
|
|
9C7J
 
 | | Crystal structure of caryolan-1-ol synthase complexed with 2-fluorofarnesyl diphosphate | | Descriptor: | (+)-caryolan-1-ol synthase, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, DIPHOSPHATE, ... | | Authors: | Prem Kumar, R, Black, B.Y, Oprian, D.D. | | Deposit date: | 2024-06-10 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Caryolan-1-ol Synthase, a Sesquiterpene Synthase Catalyzing an Initial Anti-Markovnikov Cyclization Reaction.
Biochemistry, 63, 2024
|
|
9C7I
 
 | | Crystal structure of caryolan-1-ol synthase from S. griseus with PEG molecule in the active site | | Descriptor: | (+)-caryolan-1-ol synthase, CALCIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2024-06-10 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Caryolan-1-ol Synthase, a Sesquiterpene Synthase Catalyzing an Initial Anti-Markovnikov Cyclization Reaction.
Biochemistry, 63, 2024
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9D7I
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 KN Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7H
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 KN Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7G
 
 | | BG505 DS-SOSIP.664 apo structure from the CH103 KN cryo-EM dataset | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface protein gp120, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7O
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7P
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
7COY
 
 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
7UN1
 
 | | 8-nm repeat of the human sperm tip singlet microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2FA0
 
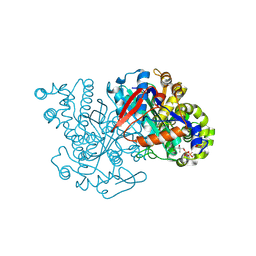 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5AJ9
 
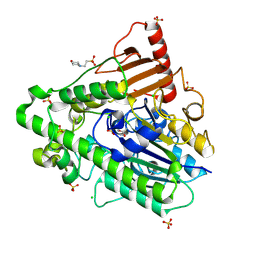 | | G7 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Fischer, G, Jonas, S, Mohammed, M.F, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1QVP
 
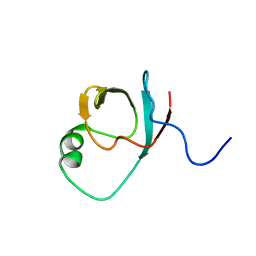 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
3IXM
 
 | |
4I64
 
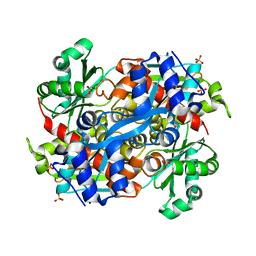 | | 3-hydroxy-3-methylglutaryl Coenzyme A reductase from Pseudomonas mevalonii, a high resolution native structure | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
3QAU
 
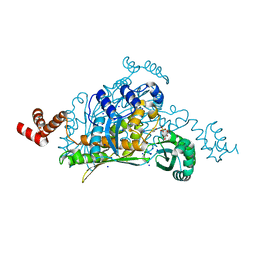 | | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, GLYCEROL, SODIUM ION, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae
To be Published
|
|
4FD6
 
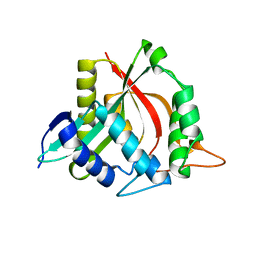 | |
3QAE
 
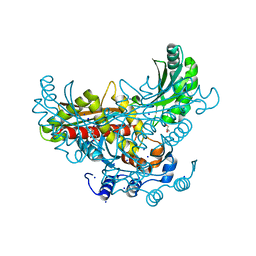 | | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae
To be Published
|
|
2OGH
 
 | | Solution structure of yeast eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Reibarkh, M, del Rio, F, Yamamoto, Y, Asano, K, Wagner, G. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Initiation Factor (eIF) 1 Carries Two Distinct eIF5-binding Faces Important for Multifactor Assembly and AUG Selection.
J.Biol.Chem., 283, 2008
|
|
