8WAS
 
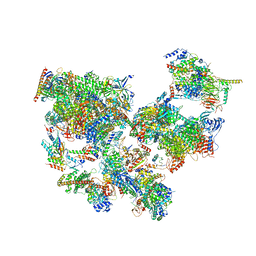 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAR
 
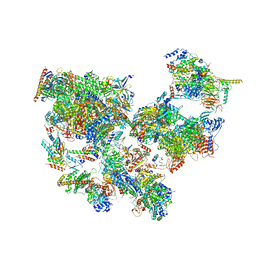 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAQ
 
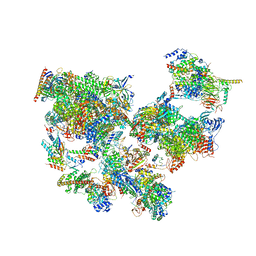 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAP
 
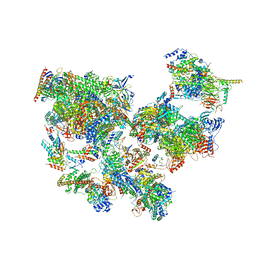 | | Structure of transcribing complex 6 (TC6), the initially transcribing complex with Pol II positioned 6nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAO
 
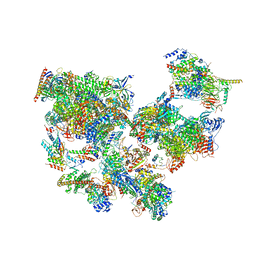 | | Structure of transcribing complex 5 (TC5), the initially transcribing complex with Pol II positioned 5nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAN
 
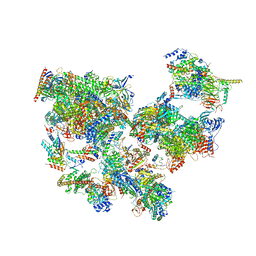 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAL
 
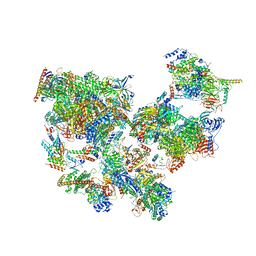 | | Structure of transcribing complex 3 (TC3), the initially transcribing complex with Pol II positioned 3nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (8.52 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAK
 
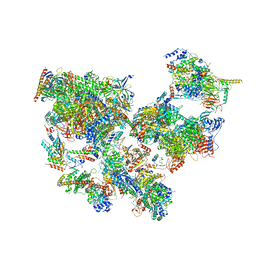 | | Structure of transcribing complex 2 (TC2), the initially transcribing complex with Pol II positioned 2nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAG
 
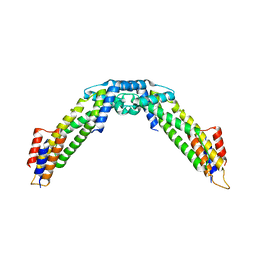 | | Crystal structure of the C-terminal fragment (residues 716-982) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Nakamura, Y, Takano, A, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
8WAF
 
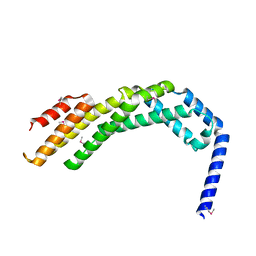 | | Crystal structure of the C-terminal fragment (residues 756-982 with the C864S mutation) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Takano, A, Nakamura, Y, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
8WAA
 
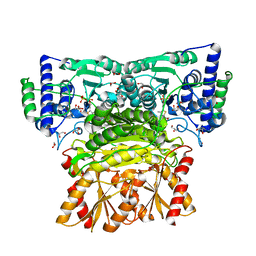 | | Human transketolase soaked with donor ketose D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA7
 
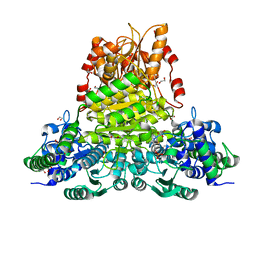 | | E.coli transketolase soaked with donor ketose D-fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA5
 
 | |
8WA3
 
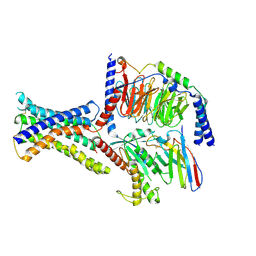 | | Cryo-EM structure of peptide free and Gs-coupled GIPR | | Descriptor: | Gastric inhibitory polypeptide receptor,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,O-antigen polymerase, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WA2
 
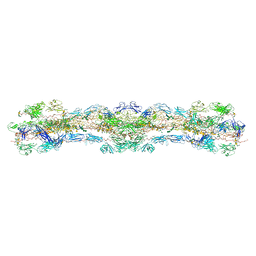 | | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, J, Tao, H, Chen, J, Pan, J, Yan, C, Yan, N. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-guided discovery of protein and glycan components in native mastigonemes.
Cell, 187, 2024
|
|
8W9Y
 
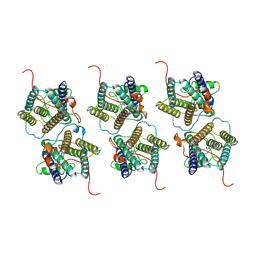 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W9J
 
 | | Crystal structure of human CLEC12A ectodomain complexed with 50C1 Fab | | Descriptor: | Anti-human CLEC12A antibody 50C1 heavy chain, Anti-human CLEC12A antibody 50C1 light chain, C-type lectin domain family 12 member A | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
8W9H
 
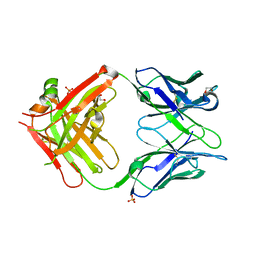 | | Crystal structure of anti-human CLEC12A antibody 50C1 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, anti-human CLEC12A antibody 50C1 Fab Heavy chain, ... | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
8W9F
 
 | |
8W9E
 
 | |
8W9D
 
 | |
8W9C
 
 | |
8W8T
 
 | | Crystal structure of human CLEC12A CRD | | Descriptor: | C-type lectin domain family 12 member A, SULFATE ION | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
8W8K
 
 | | Crystal structures of HSP90 and the compound Ganetespid in the "closed" conformation | | Descriptor: | 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(1-methyl-1H-indol-5-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-09-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
