7X0T
 
 | |
7X07
 
 | | Cryo-EM structure of human ABCD1 in the presence of C26:0 | | Descriptor: | ATP-binding cassette sub-family D member 1, hexacosanoic acid | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-02-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
7X1W
 
 | |
7XEC
 
 | |
7Y1J
 
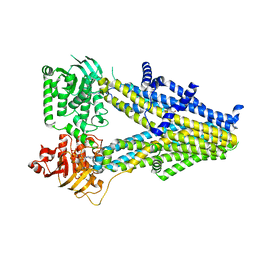 | | Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1L
 
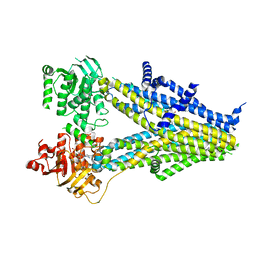 | | Structure of SUR2B in complex with Mg-ATP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1K
 
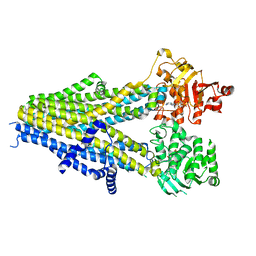 | | Structure of SUR2A in complex with Mg-ATP, Mg-ADP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1M
 
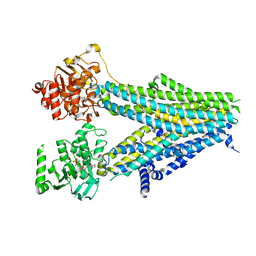 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1N
 
 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the partially occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z19
 
 | | E. coli C-P lyase bound to a single PhnK ABC domain | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z17
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z15
 
 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z16
 
 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7YRQ
 
 | | Cryo-EM structure of human Peroxisomal ABC Transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
7ZDR
 
 | |
7ZDB
 
 | |
7ZDG
 
 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDS
 
 | |
7ZDV
 
 | |
7ZDF
 
 | |
7ZDT
 
 | |
7ZDE
 
 | |
7ZEC
 
 | |
7ZDW
 
 | |
