7RD7
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
3HJB
 
 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
4U9E
 
 | |
6RND
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 15 ms timepoint | | Descriptor: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
3HHA
 
 | | Crystal structure of cathepsin L in complex with AZ12878478 | | Descriptor: | ACETATE ION, Cathepsin L1, GLYCEROL, ... | | Authors: | Asaad, N, Bethel, P.A, Coulson, M.D, Dawson, J, Ford, S.J, Gerhardt, S, Grist, M, Hamlin, G.A, James, M.J, Jones, E.V, Karoutchi, G.I, Kenny, P.W, Morley, A.D, Oldham, K, Rankine, N, Ryan, D, Wells, S.L, Wood, L, Augustin, M, Krapp, S, Simader, H, Steinbacher, S. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Dipeptidyl nitrile inhibitors of Cathepsin L.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7R7D
 
 | | Structure of human SHP2 in complex with compound 22 | | Descriptor: | 4-[6-(4-amino-4-methylpiperidin-1-yl)-1H-pyrazolo[3,4-b]pyrazin-3-yl]-3-chloro-N-methylpyridin-2-amine, TETRAETHYLENE GLYCOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6QNJ
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 4.5 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
7RP0
 
 | | Structural Snapshots of Intermediates in the Gating of a K+ Channel | | Descriptor: | DIACYL GLYCEROL, KcsA Fab chain A, KcsA Fab chain B, ... | | Authors: | Reddi, R, Matulef, K, Riederer, E.A, Valiyaveetil, F.I. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of Gating Intermediates in a K + channell.
J.Mol.Biol., 433, 2021
|
|
7R86
 
 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
6QNK
 
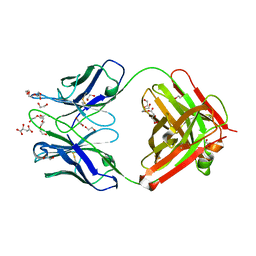 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
4UAC
 
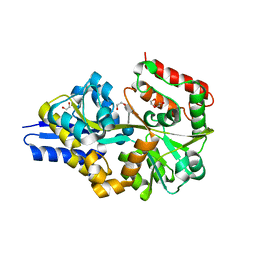 | | EUR_01830 with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Carbohydrate ABC transporter substrate-binding protein, ... | | Authors: | Koropatkin, N.M, Orlovsky, N.I. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale.
Mol.Microbiol., 95, 2015
|
|
6QND
 
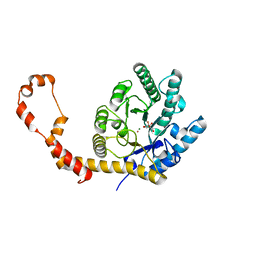 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 60 s timepoint | | Descriptor: | COBALT (II) ION, D-glucose, MAGNESIUM ION, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6QNI
 
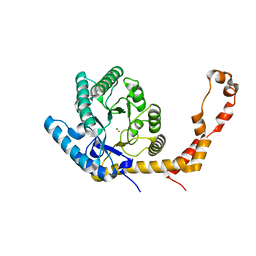 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 1.0 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
4UC2
 
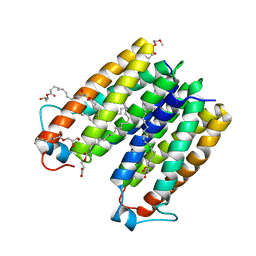 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
3II9
 
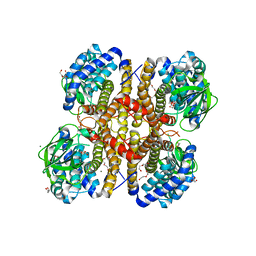 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
6IHE
 
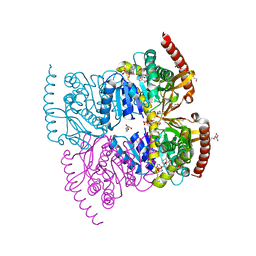 | |
6M96
 
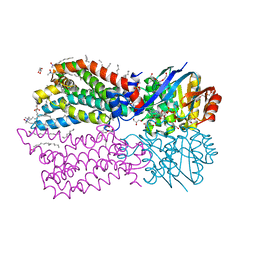 | |
6MOI
 
 | | Dimeric DARPin C_angle_R5 complex with EpoR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimeric DARPing CCR5 (C_angle_R5), ... | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C, Guo, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
3ZQA
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT C286A FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Torres-Larios, A, Munoz-Clares, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel Nadph-Cysteine Covalent Adduct Found in the Active Site of an Aldehyde Dehydrogenase.
Biochem.J., 439, 2011
|
|
3NEE
 
 | | Wild type human transthyretin (TTR) complexed with GC-1 (TTRwt:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
3NEX
 
 | |
6VJ7
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
6NAL
 
 | |
3SH3
 
 | | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii STANDL | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rangel, T.B.A, Rocha, B.A.M, Bezerra, G.A, Bezerra, M.J.B, Nascimento, K.S, Nagano, C.S, Sampaio, A.H, Assreuy, A.M.S, Gruber, K, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Biochimie, 94, 2012
|
|
6NSW
 
 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.135 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
