3TTA
 
 | |
2B5Y
 
 | |
2D0E
 
 | | Substrate assited in Oxygen Activation in Cytochrome P450 158A2 | | Descriptor: | 2-HYDROXYNAPHTHOQUINONE, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2005-08-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of active site water molecules and substrate hydroxyl groups in oxygen activation by cytochrome P450 158A2: a new mechanism of proton transfer
J.Biol.Chem., 280, 2005
|
|
3UN3
 
 | | phosphopentomutase T85Q variant soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
5U0T
 
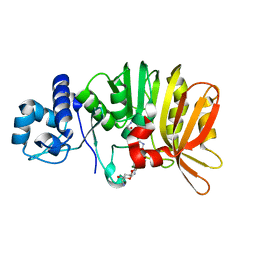 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
1HDP
 
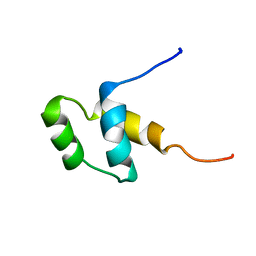 | | SOLUTION STRUCTURE OF A POU-SPECIFIC HOMEODOMAIN: 3D-NMR STUDIES OF HUMAN B-CELL TRANSCRIPTION FACTOR OCT-2 | | Descriptor: | OCT-2 POU HOMEODOMAIN | | Authors: | Sivaraja, M, Botfield, M.C, Mueller, M, Jancso, A, Weiss, M.A. | | Deposit date: | 1994-03-08 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a POU-specific homeodomain: 3D-NMR studies of human B-cell transcription factor Oct-2.
Biochemistry, 33, 1994
|
|
3TPN
 
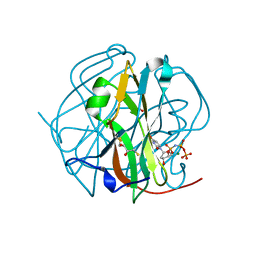 | | Crystal structure of M-PMV dUTPASE complexed with dUPNPP, substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nemeth, V, Barabas, O, Vertessy, G.B. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
To be Published
|
|
2FF7
 
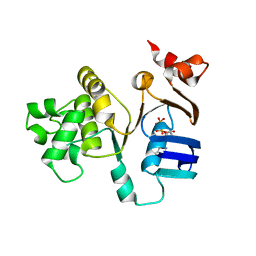 | | The ABC-ATPase of the ABC-transporter HlyB in the ADP bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-19 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
1SS1
 
 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
32C2
 
 | | STRUCTURE OF AN ACTIVITY SUPPRESSING FAB FRAGMENT TO CYTOCHROME P450 AROMATASE | | Descriptor: | IGG1 ANTIBODY 32C2 | | Authors: | Sawicki, M.W, Ng, P.C, Burkhart, B, Pletnev, V, Higashiyama, T, Osawa, Y, Ghosh, D. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an activity suppressing Fab fragment to cytochrome P450 aromatase: insights into the antibody-antigen interactions.
Mol.Immunol., 36, 1999
|
|
2DMC
 
 | | Solution structure of the 18th Filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 18th Filamin domain from human Filamin-B
To be Published
|
|
2DIA
 
 | | Solution structure of the 10th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-29 | | Release date: | 2006-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 10th filamin domain from human Filamin-B
To be Published
|
|
4EZ5
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | Cyclin-dependent kinase 6, {5-[4-(dimethylamino)piperidin-1-yl]-1H-imidazo[4,5-b]pyridin-2-yl}[2-(isoquinolin-4-yl)pyridin-4-yl]methanone | | Authors: | Chopra, R, Xu, M. | | Deposit date: | 2012-05-02 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
2E9I
 
 | | Solution structure of the N-terminal extended 20th Filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal extended 20th Filamin domain from human Filamin-B
To be Published
|
|
5U19
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6R)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
2EED
 
 | | Solution structure of the 24th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 24th filamin domain from human Filamin-B
To be Published
|
|
2HH4
 
 | | NMR structure of human insulin mutant GLY-B8-D-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | insulin A chain, insulin B chain | | Authors: | Hua, Q.X, Nakagawa, S, Hu, S.Q, Jia, W, Weiss, M.A. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Active Conformation of Insulin: Stereospecific modulation of a structural switch in the B chain.
J.Biol.Chem., 281, 2006
|
|
2HHO
 
 | | NMR structure of human insulin mutant GLY-B8-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Nakagawa, S, Hu, S.Q, Jia, W, Weiss, M.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Active Conformation of Insulin: Stereospecific modulation of a structural switch in the B chain.
J.Biol.Chem., 281, 2006
|
|
1VJD
 
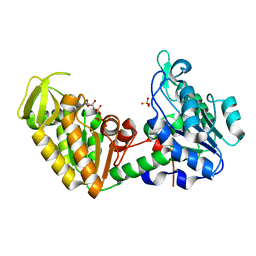 | | Structure of pig muscle PGK complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, phosphoglycerate kinase | | Authors: | Flachner, B, Kovari, Z, Varga, A, Gugolya, Z, Vonderviszt, F, Naray-Szabo, G, Vas, M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of phosphate chain mobility of MgATP in completing the 3-phosphoglycerate kinase catalytic site: binding, kinetic, and crystallographic studies with ATP and MgATP.
Biochemistry, 43, 2004
|
|
4P7L
 
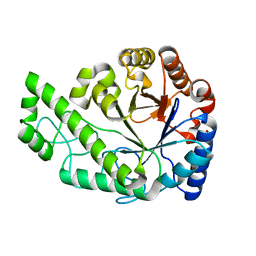 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1F0W
 
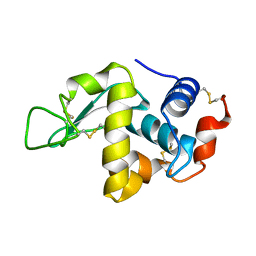 | | CRYSTAL STRUCTURE OF ORTHORHOMBIC LYSOZYME GROWN AT PH 6.5 | | Descriptor: | LYSOZYME | | Authors: | Biswal, B.K, Sukumar, N, Vijayan, M. | | Deposit date: | 2000-05-17 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydration, mobility and accessibility of lysozyme: structures of a pH 6.5 orthorhombic form and its low-humidity variant and a comparative study involving 20 crystallographically independent molecules.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4QTH
 
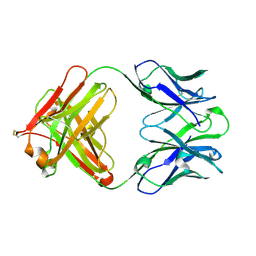 | | Crystal structure of anti-uPAR Fab 8B12 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
2FEI
 
 | | Solution structure of the second SH3 domain of Human CMS protein | | Descriptor: | CD2-associated protein | | Authors: | Yao, B, Dai, H, Jiao, Y, Wu, J, Shi, Y. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human CMS and a newly identified binding site at the C-terminus of c-Cbl
Biochim.Biophys.Acta, 1774, 2007
|
|
1WHG
 
 | | Solution structure of the CAP-Gly domain in mouse tubulin specific chaperone B | | Descriptor: | Tubulin specific chaperone B | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CAP-Gly domain in mouse tubulin specific chaperone B
To be Published
|
|
1WT0
 
 | | Mutant human ABO(H) blood group glycosyltransferase A | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Lee, H.J, Barry, C.H, Borisova, S.N, Seto, N.O.L, Zheng, R.B, Blancher, A, Evans, S.V, Palcic, M.M. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inactivity of human blood group o2 glycosyltransferase
J.Biol.Chem., 280, 2005
|
|
