7GTT
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000148a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(3,4-dihydroquinoline-1(2H)-carbothioyl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
1YXQ
 
 | | Crystal structure of actin in complex with swinholide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2005-02-22 | | Release date: | 2005-05-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of swinholide a binding to actin
Chem.Biol., 12, 2005
|
|
6KXK
 
 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
7GSR
 
 | |
7GS7
 
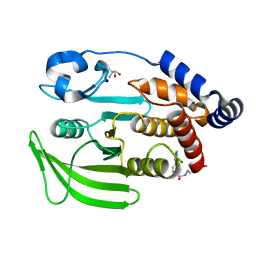 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000621a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1,2,3-thiadiazol-4-yl)phenyl ethylcarbamate, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
5DKI
 
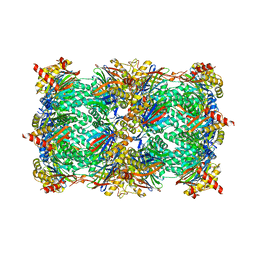 | | Yeast 20S proteasome in complex with alkyne-PI | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Cui, H, Groll, M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-28 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted Delivery of Proteasome Inhibitors to Somatostatin-Receptor-Expressing Cancer Cells by Octreotide Conjugation.
Chemmedchem, 10, 2015
|
|
157L
 
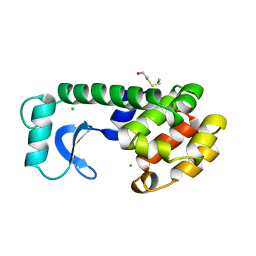 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
4HF7
 
 | |
3MG6
 
 | | Structure of yeast 20S open-gate proteasome with Compound 6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~2~-{[(1S)-6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl]acetyl}-N-{(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
4LDL
 
 | | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-[(1R)-1-hydroxy-2-{[1-(4-hydroxyphenyl)-2-methylpropan-2-yl]amino}ethyl]benzene-1,2-diol, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
1Z85
 
 | |
4HHP
 
 | | Crystal structure of triosephosphate isomerase from trypanosoma cruzi, mutant e105d | | Descriptor: | GLYCEROL, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2012-10-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
7FU8
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(2-chloro-5-pyridin-4-ylphenyl)methylamino]-5-propyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-2-({[2-chloro-5-(pyridin-4-yl)phenyl]methyl}amino)-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
4AN0
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-3 | | Descriptor: | (2S)-1-[1-(4-phenylbutanoyl)-L-prolyl]pyrrolidine-2-carbonitrile, GLYCEROL, Prolyl endopeptidase | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular dynamics, crystallography and mutagenesis studies on the substrate gating mechanism of prolyl oligopeptidase.
Biochimie, 94, 2012
|
|
7FU9
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(2-chloro-5-pyridin-4-ylphenyl)methylamino]-5-propyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-2-({[2-chloro-5-(pyridin-4-yl)phenyl]methyl}amino)-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
2ZAV
 
 | |
4GOT
 
 | |
2DA1
 
 | | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
1NO6
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | Descriptor: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
6LFW
 
 | |
175L
 
 | |
7FUO
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(2-chlorophenyl)methylamino]-5-[(2-fluoroanilino)methyl]-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8R)-2-{[(2-chlorophenyl)methyl]amino}-5-[(2-fluoroanilino)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
1O9V
 
 | | F17-aG lectin domain from Escherichia coli in complex with a selenium carbohydrate derivative | | Descriptor: | F17-AG LECTIN DOMAIN, methyl 2-acetamido-2-deoxy-1-seleno-beta-D-glucopyranoside | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1ZCN
 
 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4PTV
 
 | | Halothermothrix orenii beta-glucosidase A, thiocellobiose complex | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CESIUM ION, Glycoside hydrolase family 1, ... | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
