6BXU
 
 | |
2VAH
 
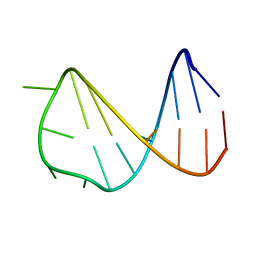 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
4APJ
 
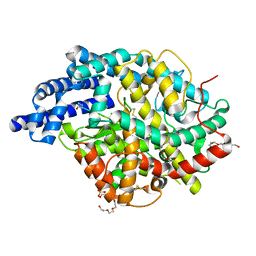 | | Human angiotensin-converting enzyme in complex with BPPb | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Schwager, S.L.U, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2012-04-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition and Regulation of Human Angiotensin-I Converting Enzyme (Ace) Activity by Natural Inhibitory Peptides.
Sci.Rep., 2, 2012
|
|
3TQA
 
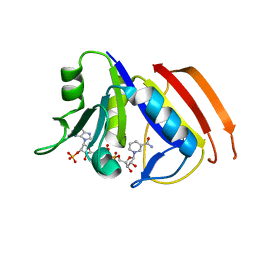 | | Structure of the dihydrofolate reductase (folA) from Coxiella burnetii in complex with NADPH | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Franklin, M.C, Cassidy, M, Hillerich, B, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4AHL
 
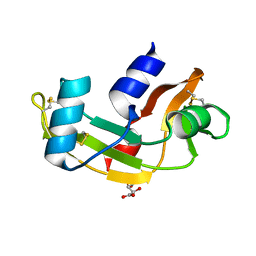 | | P112L - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | Descriptor: | ANGIOGENIN, L(+)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
479D
 
 | |
1UQT
 
 | | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | | Descriptor: | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Gibson, R.P, Tarling, C.A, Roberts, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-10-20 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The donor subsite of trehalose-6-phosphate synthase: binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution.
J. Biol. Chem., 279, 2004
|
|
3VOU
 
 | | The crystal structure of NaK-NavSulP chimera channel | | Descriptor: | COBALT (II) ION, Ion transport 2 domain protein, Voltage-gated sodium channel, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate
Nat Commun, 3, 2012
|
|
3VT5
 
 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2E,3R,5Z,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3VT7
 
 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
4CDN
 
 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
2W5N
 
 | | Native structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, GLYCEROL, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
2WFU
 
 | | Crystal structure of DILP5 variant DB | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
4B15
 
 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
4ASQ
 
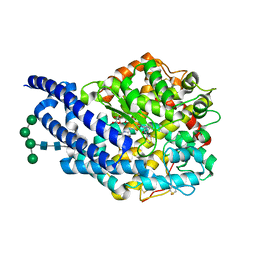 | | Crystal structure of ANCE in complex with Bradykinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ, ... | | Authors: | Akif, M, Masuyer, G, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
2W5O
 
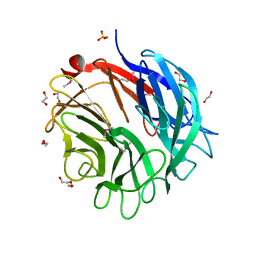 | | Complex structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum with arabinobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
3ZBV
 
 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
2W3Z
 
 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WFV
 
 | | Crystal structure of DILP5 variant C4 | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
3ZUR
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC0-A-SNAP25-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2W2D
 
 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3VT6
 
 | | Crystal structure of rat VDR-LBD with 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
