7BAO
 
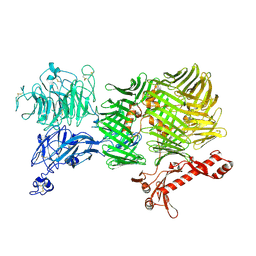 | | human Teneurin4 Mut C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meijer, D.H, Janssen, B.J.C. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Teneurin4 dimer structures reveal a calcium-stabilized compact conformation supporting homomeric trans-interactions.
Embo J., 41, 2022
|
|
5F3E
 
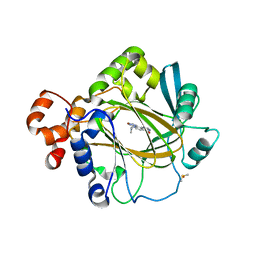 | | Crystal structure of human KDM4A in complex with compound 54a | | Descriptor: | 8-[4-[2-[4-(4-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6PUF
 
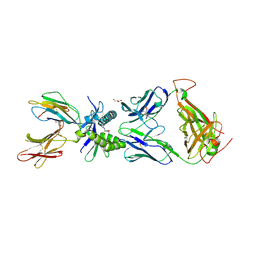 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'D-5-OP-RU | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
8D2V
 
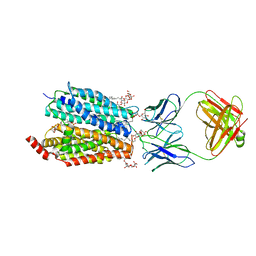 | | Zebrafish MFSD2A isoform B in inward open ligand 1B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
5AZ6
 
 | |
4XTY
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 63 with Fluorine in place of 2'OH | | Descriptor: | 2',5'-dideoxy-2'-fluoro-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.80002666 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
5EX4
 
 | | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Reichau, S, Jiao, W, Blackmore, N.J, Hutton, R.D, Parker, E.J. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with tryptophan in all three allosteric binding sites
to be published
|
|
7BAN
 
 | | human Teneurin4 Mut C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meijer, D.H, Janssen, B.J.C. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Teneurin4 dimer structures reveal a calcium-stabilized compact conformation supporting homomeric trans-interactions.
Embo J., 41, 2022
|
|
2UX8
 
 | | Crystal Structure of Sphingomonas elodea ATCC 31461 Glucose-1- phosphate uridylyltransferase in Complex with glucose-1-phosphate. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Aragao, D, Fialho, A.M, Marques, A.R, Frazao, C, Sa-Correia, I, Mitchell, E.P. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Complex of Sphingomonas Elodea Atcc 31461 Glucose-1-Phosphate Uridylyltransferase with Glucose-1-Phosphate Reveals a Novel Quaternary Structure, Unique Among Nucleoside Diphosphate-Sugar Pyrophosphorylase Members.
J.Bacteriol., 189, 2007
|
|
8D2U
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1A conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2S
 
 | | Zebrafish MFSD2A isoform B in inward open ligand bound conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
5F2P
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 3. | | Descriptor: | 2-(dimethylamino)-6-methyl-pyrido[4,3-d]pyrimidin-5-one, BRD9 | | Authors: | Nar, H, Fiegen, D, Zoephel, A, Bader, G. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
8D2W
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 2B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
8D2T
 
 | | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2X
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 3C conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8J4Z
 
 | |
6QB6
 
 | | Mcl1 in complex with a Fab | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6TP3
 
 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TO7
 
 | | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
2V8L
 
 | |
5U6E
 
 | | Crystal structure of clade A/E HIV-1 gp120 core in complex with NBD-14010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{(1S)-2-amino-1-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl]ethyl}-5-(4-chloro-3-fluorophenyl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
J. Med. Chem., 60, 2017
|
|
4XU2
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 87 with a 3'deoxy ribose | | Descriptor: | 3',5'-dideoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8500284 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
8J7D
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state
To Be Published
|
|
8J78
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state
To Be Published
|
|
