5RUZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000019685960 | | Descriptor: | 4-(1H-pyrazol-3-yl)piperidine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU3
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000161696 | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000263980802 | | Descriptor: | 4-[(2R)-2-cyclobutylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUJ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000404314 | | Descriptor: | (5-bromo-1H-indol-3-yl)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVU
 
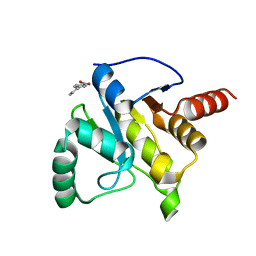 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002506130 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV0
 
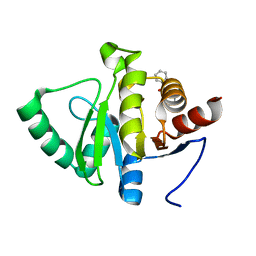 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000039994 | | Descriptor: | N-(1,3-thiazol-2-yl)benzamide, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVG
 
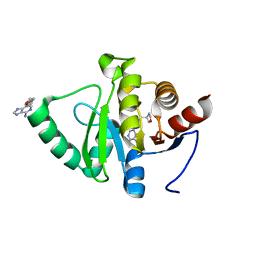 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000400552187_N3 | | Descriptor: | 3-{3-[(3S)-oxolan-3-yl]propyl}-3H-purin-6-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4JIB
 
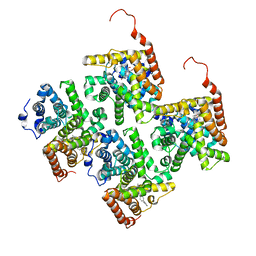 | | Crystal structure of of PDE2-inhibitor complex | | Descriptor: | 1-(2-hydroxyethyl)-3-(2-methylbutan-2-yl)-5-[4-(2-methyl-1H-imidazol-1-yl)phenyl]-6,7-dihydropyrazolo[4,3-e][1,4]diazepin-8(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of potent, selective, bioavailable phosphodiesterase 2 (PDE2) inhibitors active in an osteoarthritis pain model, Part I: Transformation of selective pyrazolodiazepinone phosphodiesterase 4 (PDE4) inhibitors into selective PDE2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7UO6
 
 | |
5C91
 
 | | NEDD4 HECT with covalently bound indole-based inhibitor | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | | Authors: | Span, I, Smith, A.T, Kathman, S, Statsyuk, A.V, Rosenzweig, A.C. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism.
J. Am. Chem. Soc., 137, 2015
|
|
3UK2
 
 | |
5DU3
 
 | | Active form of human C1-inhibitor | | Descriptor: | Plasma protease C1 inhibitor | | Authors: | Pannu, N.S, Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
1QNU
 
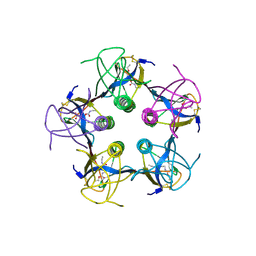 | | Shiga-Like Toxin I B Subunit Complexed with the Bridged-Starfish Inhibitor | | Descriptor: | ETHYL-CARBAMIC ACID METHYL ESTER, METHYL-CARBAMIC ACID ETHYL ESTER, Shiga toxin 1 variant B subunit, ... | | Authors: | Pannu, N.S, Hayakawa, K, Read, R.J. | | Deposit date: | 1999-10-21 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands.
Nature, 403, 2000
|
|
4HJI
 
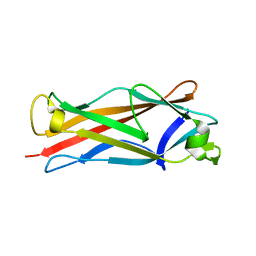 | |
4H0J
 
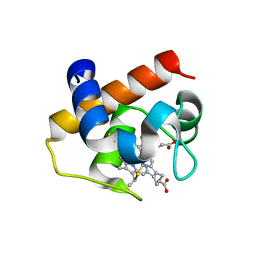 | | Mutant M58C of Nostoc sp Cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Ubbink, M, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
4H0K
 
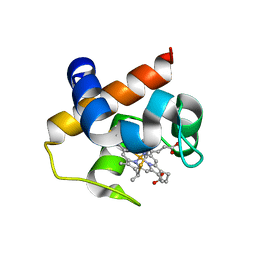 | | Mutant m58h of Nostoc sp cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Cavazzini, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
5R8L
 
 | |
4G7X
 
 | |
4G7W
 
 | |
4IWP
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
5R8P
 
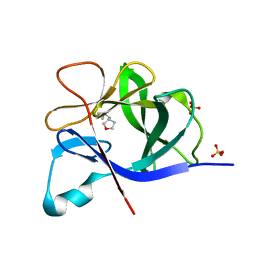 | |
4IWQ
 
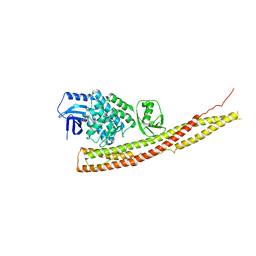 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IWO
 
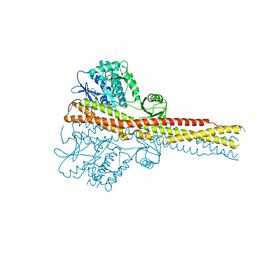 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(2-oxopyrrolidin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
7FAX
 
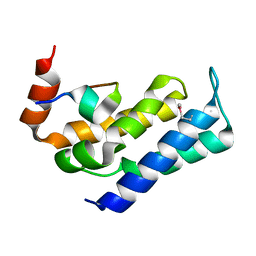 | |
7M1E
 
 | |
