6T7Y
 
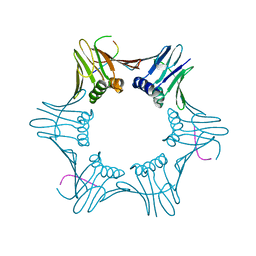 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
2FZL
 
 | |
6T7X
 
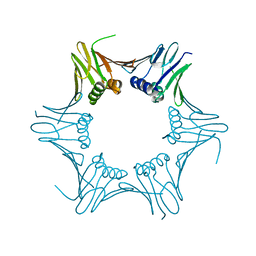 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
2KUZ
 
 | | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*TP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | Authors: | Dallmann, A, Dehmel, L, Peters, T, Muegge, C, Griesinger, C.P, Tuma, J, Ernsting, N.P. | | Deposit date: | 2010-03-03 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 2-aminopurine incorporation perturbs the dynamics and structure of DNA.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2AWA
 
 | |
2VE8
 
 | |
5WN0
 
 | | APE1 exonuclease substrate complex with a C/G match | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
5HK3
 
 | |
4LIK
 
 | |
4Z2Z
 
 | |
424D
 
 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2R7Y
 
 | |
4J4J
 
 | |
1DPN
 
 | | B-DODECAMER CGCGAA(TAF)TCGCG WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
6SJB
 
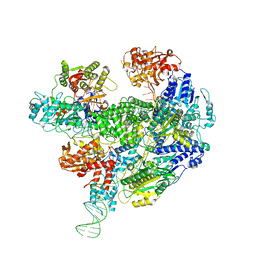 | | Cryo-EM structure of the RecBCD Chi recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
425D
 
 | | 5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7D3X
 
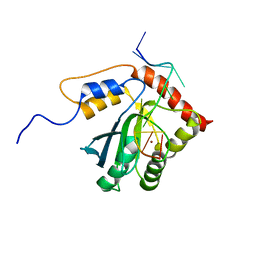 | |
2R6C
 
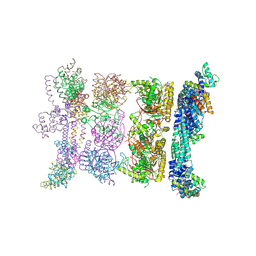 | | Crystal Form BH2 | | Descriptor: | DnaG Primase, Helicase Binding Domain, Replicative helicase | | Authors: | Bailey, S, Eliason, W.K, Steitz, T.A. | | Deposit date: | 2007-09-05 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase
Science, 318, 2007
|
|
7D3W
 
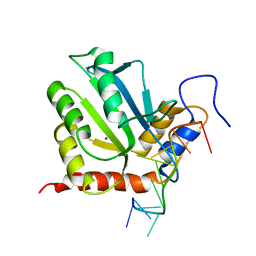 | |
2R6A
 
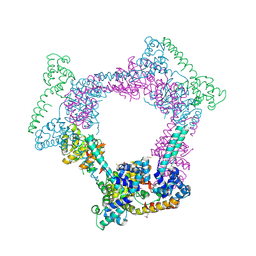 | | Crystal Form BH1 | | Descriptor: | DnaG Primase, Helicase Binding Domain, Replicative helicase, ... | | Authors: | Bailey, S, Eliason, W.K, Steitz, T.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase
Science, 318, 2007
|
|
3UC1
 
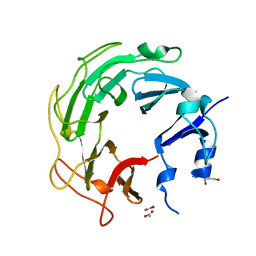 | | Mycobacterium tuberculosis gyrase type IIA topoisomerase C-terminal domain | | Descriptor: | ACETATE ION, CALCIUM ION, DNA gyrase subunit A, ... | | Authors: | Tretter, E.M, Berger, J.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms for Defining Supercoiling Set Point of DNA Gyrase Orthologs: II. THE SHAPE OF THE GyrA SUBUNIT C-TERMINAL DOMAIN (CTD) IS NOT A SOLE DETERMINANT FOR CONTROLLING SUPERCOILING EFFICIENCY.
J.Biol.Chem., 287, 2012
|
|
1OO7
 
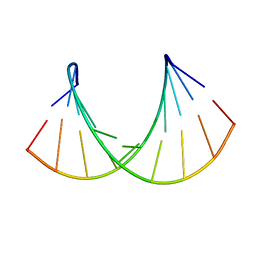 | | DNA.RNA HYBRID DUPLEX CONTAINING A 5-PROPYNE DNA STRAND AND PURINE-RICH RNA STRAND, NMR, 4 STRUCTURES | | Descriptor: | 5'-D(*GP*(5PC)P*(PDU)P*(PDU)P*(5PC)P*(PDU)P*(5PC)P*(PDU)P*(PDU)P*C)-3', 5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3' | | Authors: | Gyi, J.I, Gao, D, Conn, G.L, Trent, J.O, Brown, T, Lane, A.N. | | Deposit date: | 2003-03-03 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a DNA*RNA duplex containing 5-propynyl U and C; comparison with 5-Me modifications
Nucleic Acids Res., 31, 2003
|
|
5X9W
 
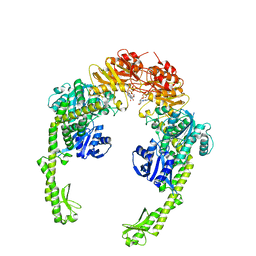 | | Mismatch Repair Protein | | Descriptor: | DNA mismatch repair protein MutS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nair, D.T, Nirwal, S. | | Deposit date: | 2017-03-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of formation of a toroid around DNA by the mismatch sensor protein.
Nucleic Acids Res., 46, 2018
|
|
2DL6
 
 | |
4DPV
 
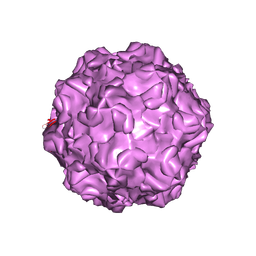 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
