7R9T
 
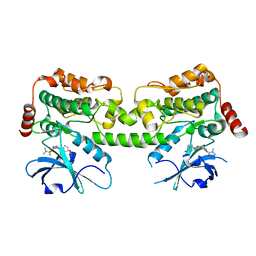 | | Crystal structure of HPK1 in complex with compound 17 | | Descriptor: | 6-amino-3-[(1S,3R)-4'-chloro-3-hydroxy-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl]-2-fluoro-N,N-dimethylbenzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
4OKF
 
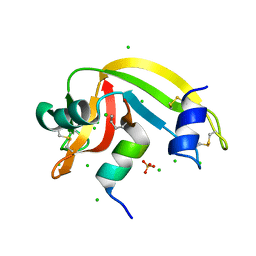 | | RNase S in complex with an artificial peptide | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2014-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | X-ray structure of a RNase S variant in complex with an artificial peptide
To be Published
|
|
7R9N
 
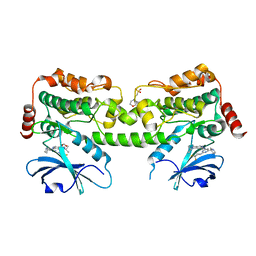 | | Crystal structure of HPK1 in complex with GNE1858 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hematopoietic progenitor kinase, ... | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6EIS
 
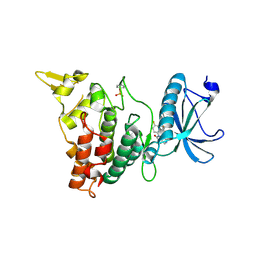 | | DYRK1A in complex with JWC-055 | | Descriptor: | 1-[4-fluoranyl-2-(trifluoromethyl)phenyl]-9-(1~{H}-pyrazol-4-yl)benzo[h][1,6]naphthyridin-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7RJI
 
 | | BthTX-II variant b, from Bothrops jararacussu venom, complexed with stearic acid | | Descriptor: | BthTX-IIb, SODIUM ION, STEARIC ACID, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | BthTX-II from Bothrops jararacussu venom has variants with different oligomeric assemblies: An example of snake venom phospholipases A 2 versatility.
Int.J.Biol.Macromol., 191, 2021
|
|
7RJZ
 
 | |
6RX7
 
 | |
6EKG
 
 | |
4OOD
 
 | | Structure of K42Y mutant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6EOC
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6ER7
 
 | | CHEMOTAXIS PROTEIN CHEY FROM Pyrococcus horikoshiI | | Descriptor: | 120aa long hypothetical chemotaxis protein (CheY) | | Authors: | Paithankar, K.S, Enderle, M.E, Wirthensohn, D, Grininger, M, Oesterhelt, D. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6RZQ
 
 | |
4ORS
 
 | | Three-dimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS5
 
 | |
6ERU
 
 | |
4OXB
 
 | |
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
4XVQ
 
 | | H-Ras Y137E | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2015-01-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding.
Faseb J., 29, 2015
|
|
6E6P
 
 | | HRAS G13D bound to GppNHp (Ha,b,c13GNP) | | Descriptor: | CALCIUM ION, GLYCEROL, GTPase HRas, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6S1U
 
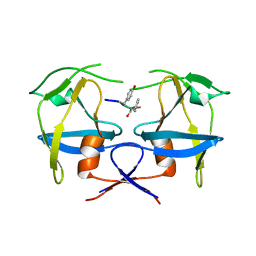 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7REH
 
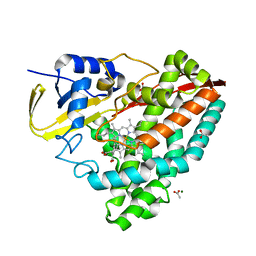 | |
6S1W
 
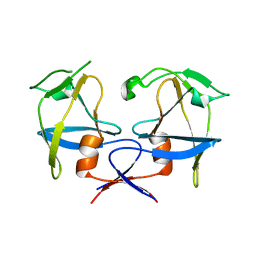 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4XX9
 
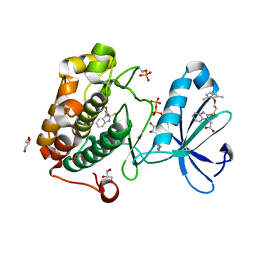 | |
4XXL
 
 | |
5TQ1
 
 | |
