3QHM
 
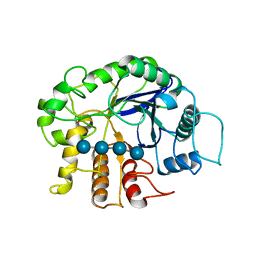 | | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
1PUU
 
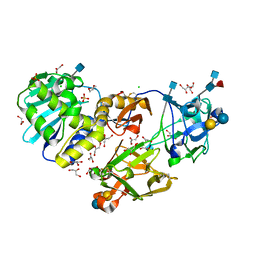 | | Mistletoe lectin I in complex with lactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krauspenhaar, R, Voelter, W, Stoeva, S, Mikhailov, A, Konareva, N, Betzel, C. | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mistletoe lectin I in complex with galactose and lactose reveals distinct sugar-binding properties
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1UMZ
 
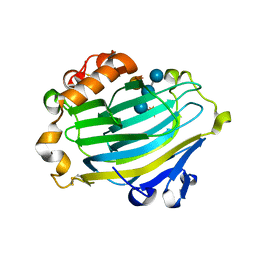 | | Xyloglucan endotransglycosylase in complex with the xyloglucan nonasaccharide XLLG. | | Descriptor: | XYLOGLUCAN ENDOTRANSGLYCOSYLASE, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A.M, Henriksson, H, Denman, S.E, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1Q25
 
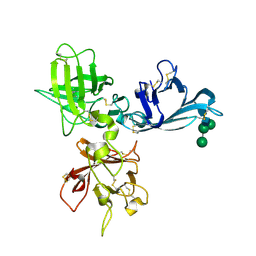 | | Crystal structure of N-terminal 3 domains of CI-MPR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2003-07-23 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of uPAR, plasminogen, and sugar-binding sites of the 300 kDa mannose 6-phosphate receptor
Embo J., 23, 2004
|
|
4FCH
 
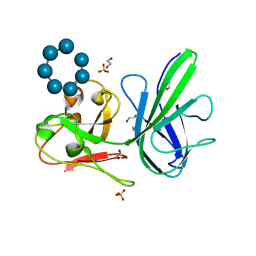 | | Crystal Structure SusE from Bacteroides thetaiotaomicron with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Outer membrane protein SusE, SULFATE ION, ... | | Authors: | Koropatkin, N.M, Cameron, E.A, Martens, E.C. | | Deposit date: | 2012-05-24 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism.
J.Biol.Chem., 287, 2012
|
|
1URX
 
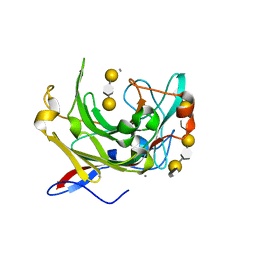 | | Crystallographic structure of beta-agarase A in complex with oligoagarose | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose, BETA-AGARASE A, ... | | Authors: | Allouch, J, Helbert, W, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Substrate Binding Sites in a Beta-Agarase Suggest a Novel Mode of Action on Double-Helical Agarose
Structure, 12, 2004
|
|
3I27
 
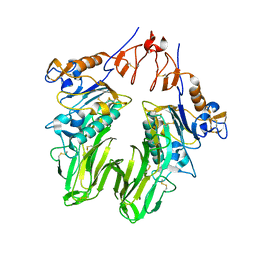 | | Structure of bovine torovirus Hemagglutinin-Esterase in complex with receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2009-06-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1UQY
 
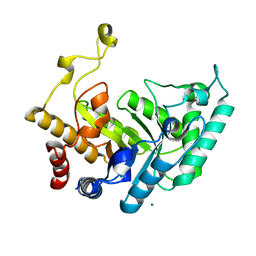 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | Descriptor: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
1HKI
 
 | | Crystal structure of human chitinase in complex with glucoallosamidin B | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
3ADZ
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with intermediate PSPP | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3VLW
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
4YGW
 
 | | RNase S in complex with stabilized S peptide | | Descriptor: | 1-hydroxypropan-2-one, Ribonuclease A C2, S-peptide: ACE-LYS-GLU-THR-ALA-ALA-HCS-LYS-PHE-GLU-HCS-GLN-HIS-MET-ASP-SER, ... | | Authors: | Assem, N, Ferreira, D, Wolan, D.W, Dawson, P.E. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2VUV
 
 | | Crystal structure of Codakine at 1.3A resolution | | Descriptor: | CALCIUM ION, CITRIC ACID, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
4FTV
 
 | |
4OYB
 
 | | Crystal Structure Analysis of the solAC | | Descriptor: | Adenylate cyclase type 10, GLYCEROL, ethyl 2-[3-[(4-azanyl-1,2,5-oxadiazol-3-yl)carbonyl]phenoxy]ethanoate | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
7E6C
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
5PB2
 
 | | Crystal Structure of Factor VIIa in complex with 2-phenyl-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-3-ol | | Descriptor: | 1-phenyl-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)-1H-pyrazol-5-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
3V9Y
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | 4-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}butanoic acid, Peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4JZS
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FV0
 
 | |
3VEV
 
 | | Glucokinase in complex with an activator and glucose | | Descriptor: | (2S)-3-cyclopentyl-N-(5-methylpyridin-2-yl)-2-[2-oxo-4-(trifluoromethyl)pyridin-1(2H)-yl]propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
5PAS
 
 | | Crystal Structure of Factor VIIa in complex with (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]-3-phenylpropanamide | | Descriptor: | (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]-3-phenylpropanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
9HEX
 
 | |
