6KXX
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
5SPT
 
 | |
5SG7
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N1=C(C(=O)C=CN1c2cccc(c2)C(F)(F)F)c3ccnn3c4c(cccc4)F, micromolar IC50=0.07 | | Descriptor: | 3-[1-(2-fluorophenyl)-1H-pyrazol-5-yl]-1-[3-(trifluoromethyl)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SPS
 
 | |
3IAP
 
 | | E. coli (lacZ) beta-galactosidase (E416Q) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
5SPQ
 
 | |
5WDD
 
 | | Crystal structure of chicken BOK | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-related ovarian killer protein | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-07-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Embryogenesis and Adult Life in the Absence of Intrinsic Apoptosis Effectors BAX, BAK, and BOK.
Cell, 173, 2018
|
|
5SDX
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cccc(n1)CCc2nc(cn2C)c3ccccc3)N4CCOCC4, micromolar IC50=3.142777 | | Descriptor: | 4-{6-[(E)-2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethen-1-yl]pyridin-2-yl}morpholine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SGY
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12nc(cn1ccc(n2)NC(c3c(cnn3CC(C)C)Cl)=O)c4ccccc4, micromolar IC50=0.018785 | | Descriptor: | 4-chloro-1-(2-methylpropyl)-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5X27
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
2WE4
 
 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | Descriptor: | CARBAMATE KINASE 1, SULFATE ION | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
5SER
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(cnn1C)C(=O)N(C)CCF)C(Nc3cc2nc(nn2cc3)c4ccccc4)=O, micromolar IC50=0.0009363 | | Descriptor: | MAGNESIUM ION, N~4~-(2-fluoroethyl)-N~4~,1-dimethyl-N~5~-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFR
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(C(=O)N(C)CC)cnn1C)C(Nc3nc2nc(cn2cc3)c4cc(ccc4)OCCF)=O, micromolar IC50=0.006128 | | Descriptor: | MAGNESIUM ION, N~4~-ethyl-N~5~-{(4S)-2-[3-(2-fluoroethoxy)phenyl]imidazo[1,2-a]pyrimidin-7-yl}-N~4~,1-dimethyl-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Gobbi, L, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
3I72
 
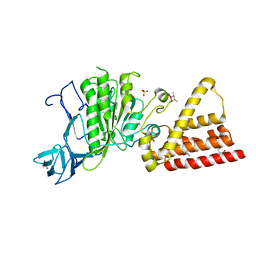 | | Structural characterization for the nucleotide binding ability of subunit A with SO4 of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
5SH6
 
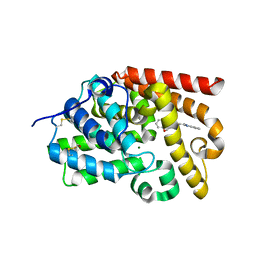 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12c(cccc1)nc(n2C)CCN4CCOc3ccccc3C4=O, micromolar IC50=2.844764 | | Descriptor: | 4-[2-(1-methyl-1H-benzimidazol-2-yl)ethyl]-3,4-dihydro-1,4-benzoxazepin-5(2H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3I7D
 
 | |
5SIJ
 
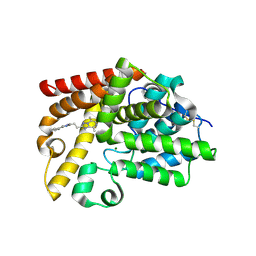 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cnc(C)c2nc(nn12)CCc3nc(cn3C)c4ccccc4)C, micromolar IC50=0.00044725 | | Descriptor: | 5,8-dimethyl-2-[2-(1-methyl-4-phenyl-imidazol-2-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SJ3
 
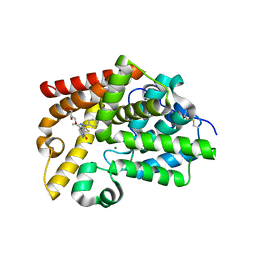 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc(s1)C(N2CCCCC2)=O)c(nn3c4ccc(Cl)cc4)C, micromolar IC50=0.143 | | Descriptor: | GLYCEROL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
6KSN
 
 | | Structure of a Zn-bound camelid single domain antibody | | Descriptor: | 1,2-ETHANEDIOL, ICab3, SODIUM ION, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2019-08-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isolation and structural characterization of a Zn2+-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria.
J.Biol.Chem., 295, 2020
|
|
5SI8
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cn(nc1C(=O)Nc3cc2nc(cn2cc3C#N)c4ccccc4)C)C(=O)N5CCOCC5, micromolar IC50=0.060125 | | Descriptor: | MAGNESIUM ION, N-[(4R)-6-cyano-2-phenylimidazo[1,2-a]pyridin-7-yl]-1-methyl-4-(morpholine-4-carbonyl)-1H-pyrazole-3-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SK0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N2(c1ccc(cn1)Cl)CC[C@H](C2)NC(c3nn(cc3c4ccncc4)C)=O, micromolar IC50=0.708457 | | Descriptor: | MAGNESIUM ION, N-[(3R)-1-(5-chloropyridin-2-yl)pyrrolidin-3-yl]-1-methyl-4-(pyridin-4-yl)-1H-pyrazole-3-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SE3
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(cc1nc(nn1cc2)c3ccccc3)NC(c4ccnc(c4)Cl)=O, micromolar IC50=0.045025 | | Descriptor: | 2-chloro-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]pyridine-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6ADT
 
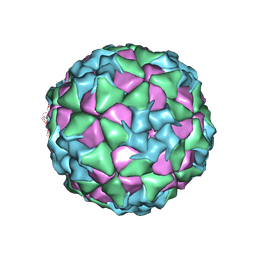 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5SEG
 
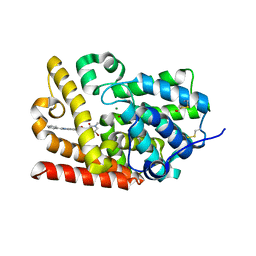 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(c(C(=O)NC1COC1)cc3c(c2)nc([nH]3)c4ccccc4)NC(C5=CC(=CN(C5=O)C)Br)=O, micromolar IC50=0.006706 | | Descriptor: | 5-[(5-bromo-1-methyl-2-oxo-1,2-dihydropyridine-3-carbonyl)amino]-N-(oxetan-3-yl)-2-phenyl-1H-benzimidazole-6-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Bleicher, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4Y2C
 
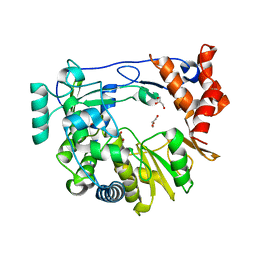 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
