5TUL
 
 | |
5TUK
 
 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUE
 
 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
1A2N
 
 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
4HP3
 
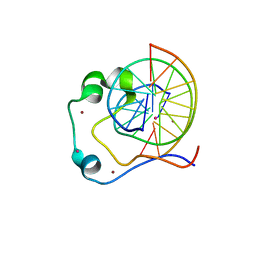 | | Crystal structure of Tet3 in complex with a CpG dsDNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), LOC100036628 protein, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
4HP1
 
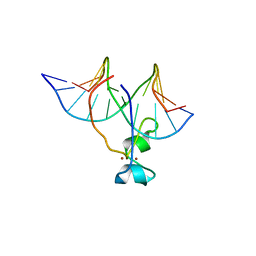 | | Crystal structure of Tet3 in complex with a non-CpG dsDNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*C)-3'), LOC100036628 protein, ZINC ION | | Authors: | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
5QGC
 
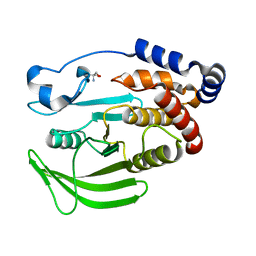 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000650a | | Descriptor: | (6aS,12aR)-3-methoxy-6a,10,11,12a-tetrahydro-6H,7H,9H-[1]benzopyrano[4,3-c]pyrazolo[1,2-a]pyrazol-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFV
 
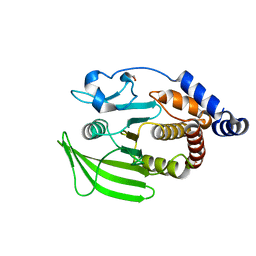 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000491a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[(1S,2S,3S,4R)-3-hydroxy-1,2,3,4-tetrahydro-1,4-epoxynaphthalen-2-yl]cyclobutanecarboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QDU
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000466a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N},~{N},5,6-tetramethylthieno[2,3-d]pyrimidin-4-amine | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QGF
 
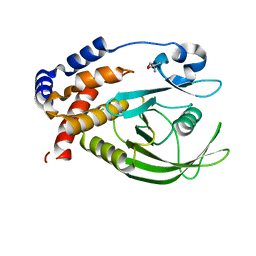 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000539a | | Descriptor: | 1-[(3S,3aS,8bS)-7-chloro-3-(hydroxymethyl)-2,3,3a,8b-tetrahydro-1H-[1]benzofuro[3,2-b]pyrrol-1-yl]ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QDT
 
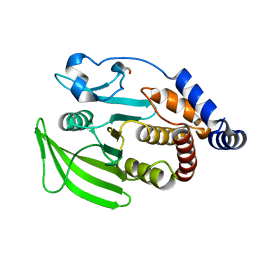 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000475a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}-(1-propyl-1,2,3,4-tetrazol-5-yl)furan-2-carboxamide | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFP
 
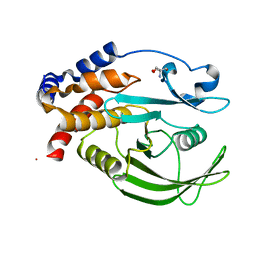 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFF
 
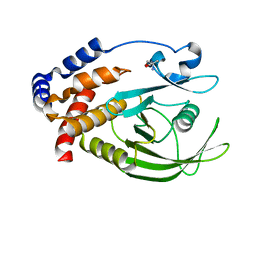 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000515a | | Descriptor: | 1-[(3S,3aS,8bS)-5-fluoro-3-(hydroxymethyl)-2,3,3a,8b-tetrahydro-1H-[1]benzofuro[3,2-b]pyrrol-1-yl]ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFS
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QG3
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000662a | | Descriptor: | (6aR,12aR)-3-methoxy-6a,10,11,12a-tetrahydro-6H,7H,9H-[1]benzopyrano[4,3-c]pyrazolo[1,2-a]pyrazol-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFQ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000491a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[(1S,2S,3S,4R)-3-hydroxy-1,2,3,4-tetrahydro-1,4-epoxynaphthalen-2-yl]cyclobutanecarboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1ICT
 
 | | MONOCLINIC FORM OF HUMAN TRANSTHYRETIN COMPLEXED WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Cody, V. | | Deposit date: | 2001-04-02 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a new polymorphic monoclinic form of human transthyretin at 3 A resolution reveals a mixed complex between unliganded and T4-bound tetramers of TTR.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7ZNY
 
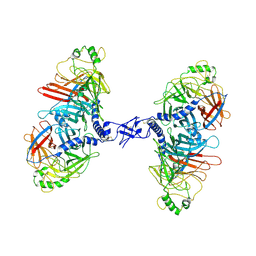 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ZA3
 
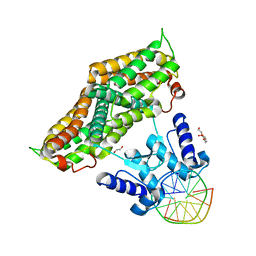 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
5M2L
 
 | |
1SFK
 
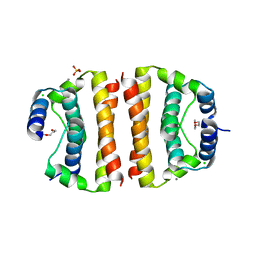 | | Core (C) protein from West Nile Virus, subtype Kunjin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | Authors: | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
1EZW
 
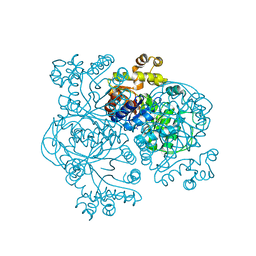 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
5Z20
 
 | | The ternary structure of D-lactate dehydrogenase from Pseudomonas aeruginosa with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase (Fermentative), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
1UX1
 
 | | Bacillus subtilis cytidine deaminase with a Cys53His and an Arg56Gln substitution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ... | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UWZ
 
 | | Bacillus subtilis cytidine deaminase with an Arg56 - Ala substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
