1IOW
 
 | | COMPLEX OF Y216F D-ALA:D-ALA LIGASE WITH ADP AND A PHOSPHORYL PHOSPHINATE | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALA:D-ALA LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-alanine:D-alanine ligase: phosphonate and phosphinate intermediates with wild type and the Y216F mutant.
Biochemistry, 36, 1997
|
|
1CFA
 
 | | SOLUTION STRUCTURE OF A SEMI-SYNTHETIC C5A RECEPTOR ANTAGONIST AT PH 5.2, 303K, NMR, 20 STRUCTURES | | Descriptor: | COMPLEMENT 5A SEMI-SYNTHETIC ANTAGONIST, SYNTHETIC N-TERMINAL TAIL | | Authors: | Zhang, X, Boyar, W, Galakatos, N, Gonnella, N.C. | | Deposit date: | 1996-09-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding.
Protein Sci., 6, 1997
|
|
1VDC
 
 | | STRUCTURE OF NADPH DEPENDENT THIOREDOXIN REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DEPENDENT THIOREDOXIN REDUCTASE, SULFATE ION | | Authors: | Dai, S, Eklund, H. | | Deposit date: | 1996-09-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 A resolution.
J.Mol.Biol., 264, 1996
|
|
1YBV
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
219L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
210L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
212L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
211L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
209L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
218L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
215L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
213L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
214L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
1NEU
 
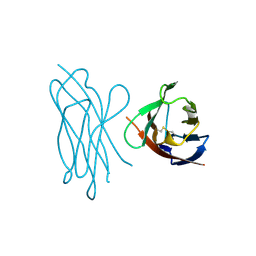 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1ZDI
 
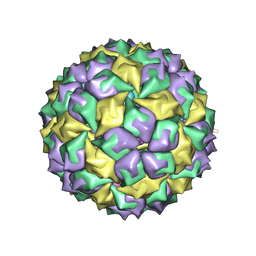 | | RNA BACTERIOPHAGE MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (RNA BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1ZDH
 
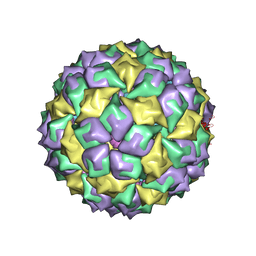 | | MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1DJG
 
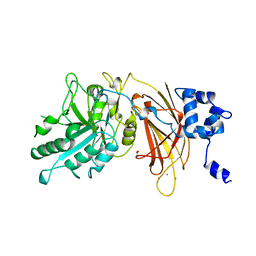 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH LANTHANUM | | Descriptor: | ACETATE ION, LANTHANUM (III) ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1NGS
 
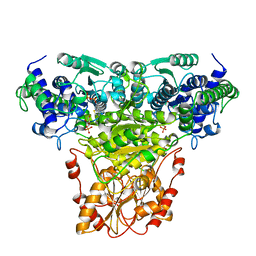 | | COMPLEX OF TRANSKETOLASE WITH THIAMIN DIPHOSPHATE, CA2+ AND ACCEPTOR SUBSTRATE ERYTHROSE-4-PHOSPHATE | | Descriptor: | CALCIUM ION, ERYTHOSE-4-PHOSPHATE, THIAMINE DIPHOSPHATE, ... | | Authors: | Nilsson, U, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis.
J.Biol.Chem., 272, 1997
|
|
1IYV
 
 | |
1IYU
 
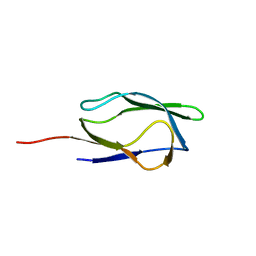 | |
1IBE
 
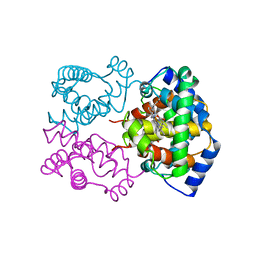 | |
1DJI
 
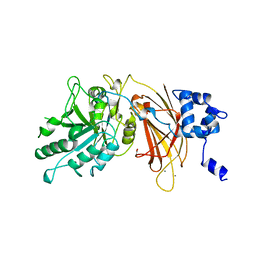 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH CALCIUM | | Descriptor: | ACETATE ION, CALCIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1DJH
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH BARIUM | | Descriptor: | ACETATE ION, BARIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1KCW
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN CERULOPLASMIN AT 3.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CERULOPLASMIN, COPPER (II) ION, ... | | Authors: | Card, G.L, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of human serum ceruloplasmin at 3.1 angstrom: Nature of the copper centres.
J.Biol.Inorg.Chem., 1, 1996
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
