5KWA
 
 | |
5KZF
 
 | |
5V8F
 
 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VC7
 
 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5VCA
 
 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | Descriptor: | VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5UIE
 
 | | Vps4-Vta1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DOA4-independent degradation protein 4, ... | | Authors: | Monroe, N, Shen, P, Han, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-12 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase.
Elife, 6, 2017
|
|
5WYJ
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WYK
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2017-05-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WVK
 
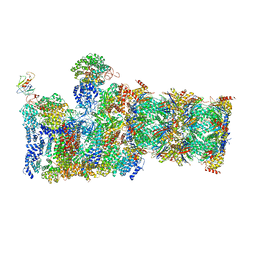 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5WVI
 
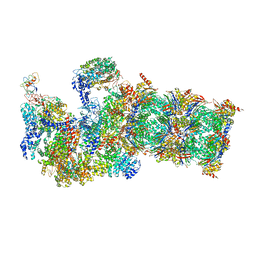 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5LN3
 
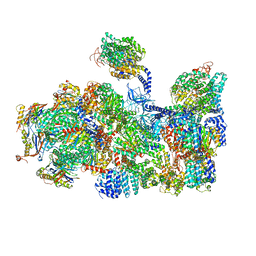 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
5MPA
 
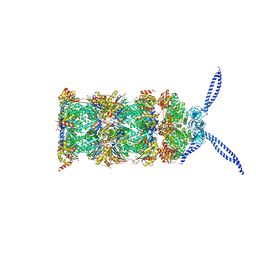 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MPC
 
 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MP9
 
 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MPB
 
 | | 26S proteasome in presence of AMP-PNP (s3) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | Authors: | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | Deposit date: | 2016-11-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (7.086 Å) | | Cite: | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
5UJM
 
 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5UJ7
 
 | | Structure of the active form of human Origin Recognition Complex ATPase motor module, complex subunitS 1, 4, 5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, Elkayam, E, On, K.F, Joshua-Tor, L. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5JI3
 
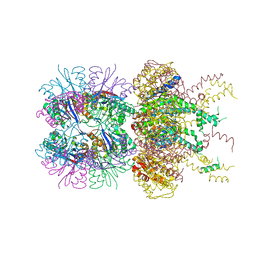 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5D4W
 
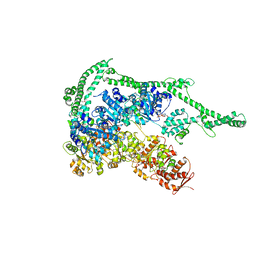 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
5JI2
 
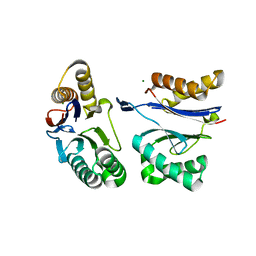 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5EQT
 
 | |
5IFW
 
 | |
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
5T0I
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
