9G7J
 
 | |
1I3N
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
9G0D
 
 | | Structure of human Mical1 bMERB_V978A domain:Rab10 complex. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-10, ... | | Authors: | Rai, A, Goody, R.S. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural insights into the auto-inhibited state of Mical1 and its activation by Rab8
Elife, 2024
|
|
9G0C
 
 | |
1IGZ
 
 | | Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
1IQY
 
 | | CRYSTAL STRUCTURE OF NICKEL-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | AMINE OXIDASE, NICKEL (II) ION | | Authors: | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2001-08-28 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
9HCJ
 
 | | D. discoideum nuclear pore complex | | Descriptor: | ELYS-like domain-containing protein, NDC1, Nuclear pore complex protein, ... | | Authors: | Obarska-Kosinska, A, Hoffmann, P.C, Beck, M. | | Deposit date: | 2024-11-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Nuclear pore permeability and fluid flow are modulated by its dilation state.
Mol.Cell, 85, 2025
|
|
9I81
 
 | | SARS-CoV-2 RdRp bound to a stack of three HeE1-2Tyr molecules | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kabinger, F, Doze, V, Schmitzova, J, Lidschreiber, M, Dienemann, C, Cramer, P. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of SARS-CoV-2 polymerase inhibition by nonnucleoside inhibitor HeE1-2Tyr.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HNF
 
 | | Beta-keto acid cleavage enzyme from Paracoccus denitrificans with bound acetoacetate and acetyl-CoA | | Descriptor: | 3-keto-5-aminohexanoate cleavage protein, ACETOACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Marchal, D.G, Zarzycki, J, Erb, T.J. | | Deposit date: | 2024-12-10 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and implementation of aerobic and ambient CO 2 -reduction as an entry-point for enhanced carbon fixation.
Nat Commun, 16, 2025
|
|
1GU6
 
 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
9I2A
 
 | |
8PSL
 
 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the ketosynthase domain (FASx sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS2
 
 | | Asymmetric unit of the yeast fatty acid synthase with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSK
 
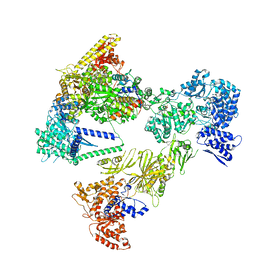 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosynthase domain (FASx sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PRV
 
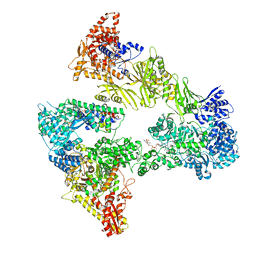 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosreductase domain (FASamn sample) | | Descriptor: | COENZYME A, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSA
 
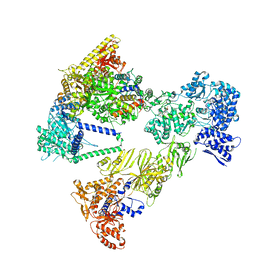 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the ketosynthase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSJ
 
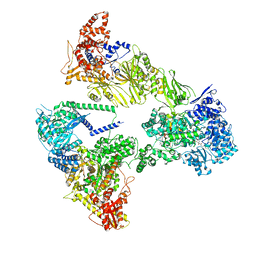 | | Asymmetric unit of the yeast fatty acid synthase in the semi rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSP
 
 | | Asymmetric unit of the yeast fatty acid synthase in rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS8
 
 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS9
 
 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosynthase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS1
 
 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosynthase domain (FASamn sample) | | Descriptor: | COENZYME A, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSG
 
 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSF
 
 | | Asymmetric unit of the yeast fatty acid synthase in non-rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
9H1P
 
 | |
1I3L
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
