4AY2
 
 | | Capturing 5' tri-phosphorylated RNA duplex by RIG-I | | Descriptor: | 5'-R-PPP(GP*GP*CP*GP*CP*GP*GP*CP*UP*UP*CP*GP*GP*CP *CP*GP*CP*GP*CP*C)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-06-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor Rig-I.
Structure, 20, 2012
|
|
1DFU
 
 | |
1S68
 
 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
1UUD
 
 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
2MQV
 
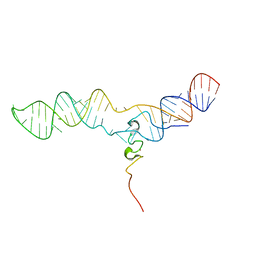 | |
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | Descriptor: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
1EXY
 
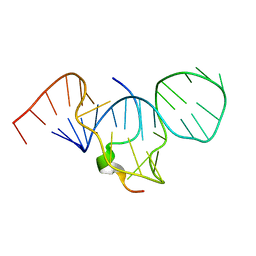 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1HMH
 
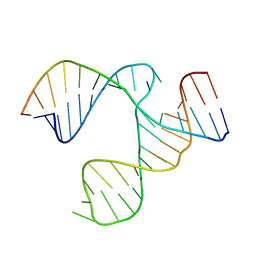 | |
1EIZ
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | FTSJ, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
7DTK
 
 | |
7DTJ
 
 | |
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
7BAI
 
 | |
7BAH
 
 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the RNA sensor RIG-I controls immune tolerance to mitochondrial RNA
To Be Published
|
|
2LI8
 
 | |
1EBR
 
 | |
1EBS
 
 | |
1EBQ
 
 | |
4X2Y
 
 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
4X2X
 
 | |
3IGI
 
 | | Tertiary Architecture of the Oceanobacillus Iheyensis Group II Intron | | Descriptor: | 5'-R(*CP*GP*CP*UP*CP*UP*AP*CP*UP*CP*UP*AP*U)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Keating, K.S, Fedorova, O, Rajashankar, K, Wang, J, Pyle, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | Tertiary architecture of the Oceanobacillus iheyensis group II intron.
Rna, 16, 2010
|
|
1I7J
 
 | | CRYSTAL STRUCTURE OF 2'-O-ME(CGCGCG)2: AN RNA DUPLEX AT 1.19 A RESOLUTION. 2-METHYL-2,4-PENTANEDIOL AND MAGNESIUM BINDING. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3', MAGNESIUM ION | | Authors: | Adamiak, D.A, Rypniewski, W.R, Milecki, J, Adamiak, R.W. | | Deposit date: | 2001-03-09 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The 1.19 A X-ray structure of 2'-O-Me(CGCGCG)(2) duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove.
Nucleic Acids Res., 29, 2001
|
|
2N3G
 
 | |
2N3H
 
 | |
