1ROO
 
 | | NMR SOLUTION STRUCTURE OF SHK TOXIN, NMR, 20 STRUCTURES | | Descriptor: | SHK TOXIN | | Authors: | Tudor, J.E, Pallaghy, P.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone.
Nat.Struct.Biol., 3, 1996
|
|
4BS2
 
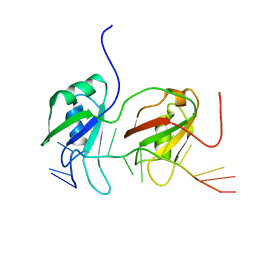 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
1NGU
 
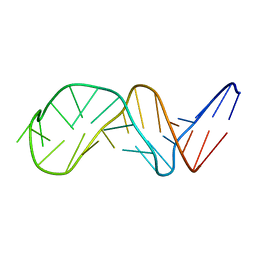 | | NMR Structure of Putative 3'Terminator for B. Anthracis pagA Gene Noncoding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*CP*CP*TP*TP*GP*TP*AP*TP*TP*TP*CP*TP*TP*AP*CP*AP*AP*AP*AP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
1NGO
 
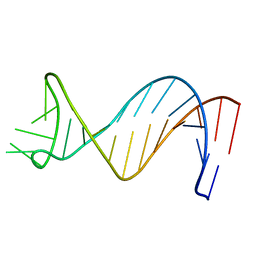 | | NMR Structure of Putative 3' Terminator for B. Anthracis pagA Gene Coding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*TP*TP*GP*TP*AP*AP*GP*AP*AP*AP*TP*AP*CP*AP*AP*GP*GP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
1OQ2
 
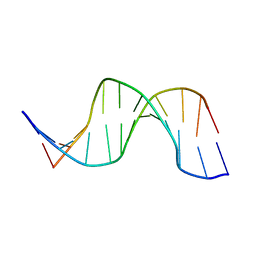 | | NMR structure of hemimethylated GATC site | | Descriptor: | 5'-D(*CP*GP*CP*AP*GP*(6MA)P*TP*CP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3' | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-07 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
1HFF
 
 | | NMR solution structures of the vMIP-II 1-10 peptide from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1OPQ
 
 | | NMR structure of unmethylated GATC site | | Descriptor: | 5'-D(*CP*GP*CP*AP*GP*AP*TP*CP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3' | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
1SKP
 
 | | NMR STRUCTURE OF D(GCATATGATAG)(DOT)D(CTATCATATGC): A CONSENSUS SEQUENCE FOR PROMOTERS RECOGNIZED BY SIGMA-K RNA POLYMERASE, 4 STRUCTURES | | Descriptor: | SIGMA-K RNA POLYMERASE CONSENSUS SEQUENCE | | Authors: | Tonelli, M, Ragg, E, Bianucci, A.M, Lesiak, K, James, T.L. | | Deposit date: | 1998-05-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of d(GCATATGATAG). d(CTATCATATGC): a consensus sequence for promoters recognized by sigma K RNA polymerase.
Biochemistry, 37, 1998
|
|
1HFG
 
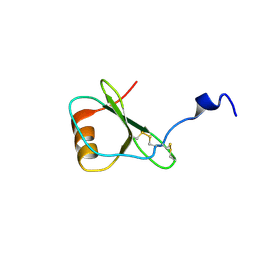 | | NMR solution structure of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus (minimized average structure). | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2001-01-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1MXJ
 
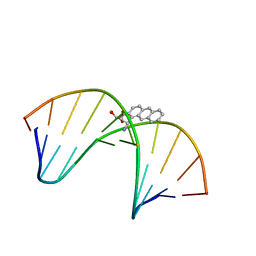 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | Descriptor: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | Authors: | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
1IMO
 
 | |
1KMF
 
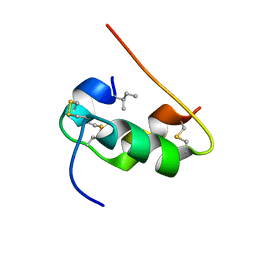 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1IBX
 
 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I17
 
 | | NMR STRUCTURE OF MOUSE DOPPEL 51-157 | | Descriptor: | PRION-LIKE PROTEIN | | Authors: | Mo, H, Moore, R.C, Cohen, F.E, Westaway, D, Prusiner, S.B, Wright, P.E, Dyson, H.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two different neurodegenerative diseases caused by proteins with similar structures.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2Z2G
 
 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2Z2H
 
 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
1NYF
 
 | |
1G4D
 
 | |
1G7O
 
 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
1GL5
 
 | |
1NYG
 
 | |
1F2R
 
 | | NMR STRUCTURE OF THE HETERODIMERIC COMPLEX BETWEEN CAD DOMAINS OF CAD AND ICAD | | Descriptor: | CASPASE-ACTIVATED DNASE, INHIBITOR OF CASPASE-ACTIVATED DNASE | | Authors: | Otomo, T, Sakahira, H, Uegaki, K, Nagata, S, Yamazaki, T. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the heterodimeric complex between CAD domains of CAD and ICAD.
Nat.Struct.Biol., 7, 2000
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2OVN
 
 | |
