1DOS
 
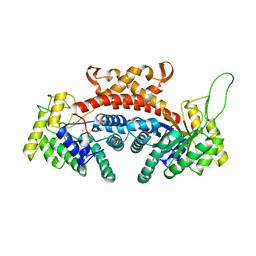 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | Descriptor: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | Authors: | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | Deposit date: | 1996-06-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
1BUF
 
 | |
1AD3
 
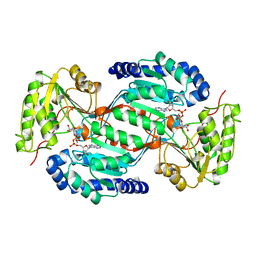 | |
1OBS
 
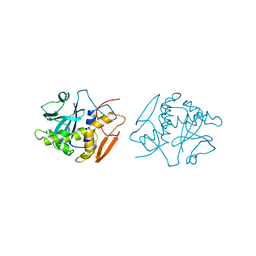 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OKN
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1OKL
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKL INHIBITOR 5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONAMIDE | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Elbaum, D, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected binding mode of the sulfonamide fluorophore 5-dimethylamino-1-naphthalene sulfonamide to human carbonic anhydrase II. Implications for the development of a zinc biosensor.
J.Biol.Chem., 271, 1996
|
|
1OKM
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKM INHIBITOR 4-SULFONAMIDE-[1-(4-AMINOBUTANE)]BENZAMIDE | | Descriptor: | 4-SULFONAMIDE-[1-(4-AMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1LSQ
 
 | | RIBONUCLEASE A WITH ASN 67 REPLACED BY A BETA-ASPARTYL RESIDUE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Esposito, L, Sica, F, Vitagliano, L, Zagari, A, Mazzarella, L. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deamidation in proteins: the crystal structure of bovine pancreatic ribonuclease with an isoaspartyl residue at position 67.
J.Mol.Biol., 257, 1996
|
|
1GGG
 
 | |
1CGH
 
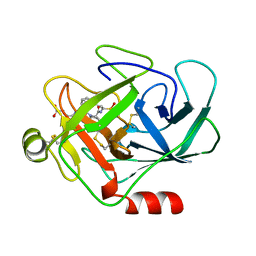 | | Human cathepsin G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-{(1R)-1-[(S)-hydroxy(oxido)phosphanyl]-2-phenylethyl}-L-prolinamide | | Authors: | Hof, P, Bode, W. | | Deposit date: | 1996-06-26 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(OPh)2: a Janus-faced proteinase with two opposite specificities.
EMBO J., 15, 1996
|
|
1UQG
 
 | |
1UQF
 
 | |
1UQB
 
 | |
1UQA
 
 | |
302D
 
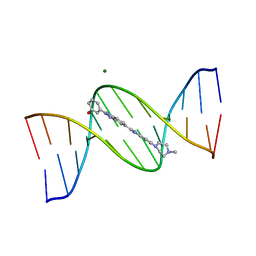 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL IN' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
1UQD
 
 | |
1UQC
 
 | |
303D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
1UQE
 
 | |
1IRV
 
 | |
2MYS
 
 | |
1IRW
 
 | |
1DRW
 
 | | ESCHERICHIA COLI DHPR/NHDH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE PURIN-6-OL-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1DRU
 
 | | ESCHERICHIA COLI DHPR/NADH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1BDD
 
 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
