5DQO
 
 | |
2K0U
 
 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
6QLC
 
 | | The ssDNA-binding RNA polymerase cofactor Drc from Pseudomonas phage LUZ7 | | Descriptor: | PHOSPHATE ION, ssDNA binding RNA Polymerase cofactor | | Authors: | De Zitter, E, Boon, M, De Smet, J, Lavigne, R, Van Meervelt, L. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 'Drc', a structurally novel ssDNA-binding transcription regulator of N4-related bacterial viruses.
Nucleic Acids Res., 48, 2020
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
2K0T
 
 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-14 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
4CC6
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
5I7M
 
 | |
3O1R
 
 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-21 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
2MIW
 
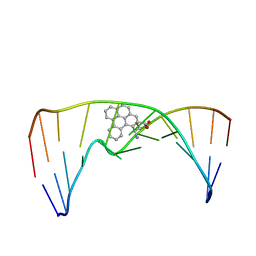 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIV
 
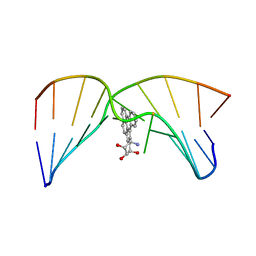 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2LJ6
 
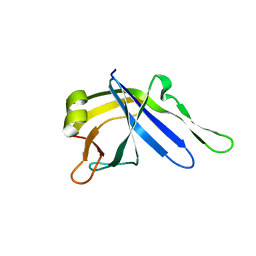 | |
8AMU
 
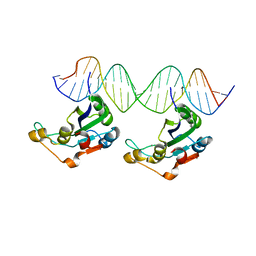 | | RepB pMV158 OBD domain bound to DDR region | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Amodio, J, Machon, C, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
6I4N
 
 | | Dodecamer DNA containing the synthetic base pair P-Z in complex with a pyrrole-imidazole polyamide | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-D(*CP*GP*AP*TP*(DP)P*TP*AP*(DZ)P*AP*TP*CP*G)-3') | | Authors: | Padroni, G, Parkison, J, Burley, G.A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
6KDL
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.274 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDP
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, FORMIC ACID, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6S7D
 
 | | Self-complementary duplex DNA containing an internucleoside phosphoroselenolate | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*GP*(XCI)P*CP*CP*CP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Conlon, P.F, Steinhogl, J, Vyle, J.S, Hall, J.P. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solid-phase synthesis and structural characterisation of phosphoroselenolate-modified DNA: a backbone analogue which does not impose conformational bias and facilitates SAD X-ray crystallography.
Chem Sci, 10, 2019
|
|
2NQJ
 
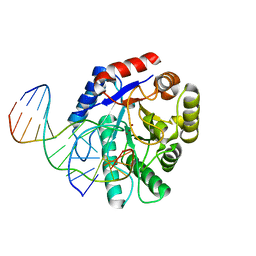 | | Crystal structure of Escherichia coli endonuclease IV (Endo IV) E261Q mutant bound to damaged DNA | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', Endonuclease 4, ... | | Authors: | Garcin-Hosfield, E.D, Hosfield, D.J, Tainer, J.A. | | Deposit date: | 2006-10-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | DNA apurinic-apyrimidinic site binding and excision by endonuclease IV.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2NQ9
 
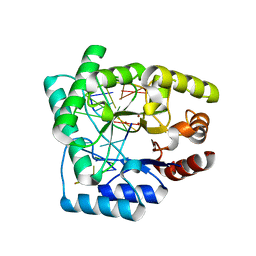 | | High resolution crystal structure of Escherichia coli endonuclease IV (Endo IV) Y72A mutant bound to damaged DNA | | Descriptor: | 5'-D(*AP*TP*AP*TP*CP*T)-3', 5'-D(*AP*TP*CP*TP*GP*AP*AP*GP*TP*AP*T)-3', 5'-D(P*(3DR)P*AP*GP*AP*T)-3', ... | | Authors: | Garcin-Hosfield, E.D, Hosfield, D.J, Tainer, J.A. | | Deposit date: | 2006-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA apurinic-apyrimidinic site binding and excision by endonuclease IV.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5CIU
 
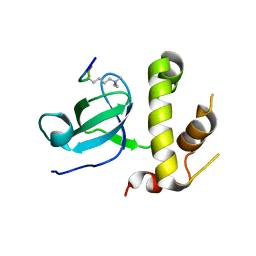 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | Authors: | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
3GOE
 
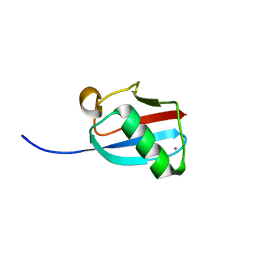 | | Molecular Mimicry of SUMO promotes DNA repair | | Descriptor: | CALCIUM ION, DNA repair protein rad60 | | Authors: | Perry, J.J.P. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular mimicry of SUMO promotes DNA repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2O8F
 
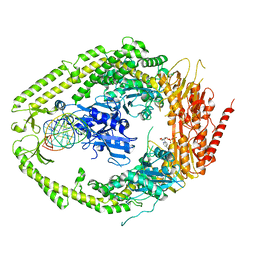 | | human MutSalpha (MSH2/MSH6) bound to DNA with a single base T insert | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*CP*GP*TP*C)-3', 5'-D(*GP*AP*CP*GP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2LYG
 
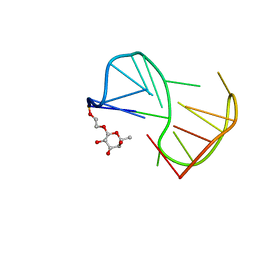 | | Fuc_TBA | | Descriptor: | 2-hydroxyethyl 6-deoxy-beta-L-galactopyranoside, DNA (5'-D(P*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avio, A, Eritja, R, Gonzalez-Ibaez, C, Morales, J. | | Deposit date: | 2012-09-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbohydrate-DNA interactions at G-quadruplexes: folding and stability changes by attaching sugars at the 5'-end.
Chemistry, 19, 2013
|
|
5IWD
 
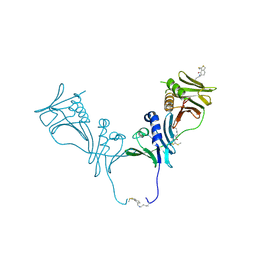 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
