2VI6
 
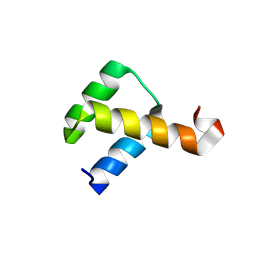 | | Crystal Structure of the Nanog Homeodomain | | Descriptor: | HOMEOBOX PROTEIN NANOG | | Authors: | Jauch, R, Ng, C.K.L, Saitakendu, K.S, Stevens, R.C, Kolatkar, P.R. | | Deposit date: | 2007-11-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog.
J.Mol.Biol., 376, 2008
|
|
8QKV
 
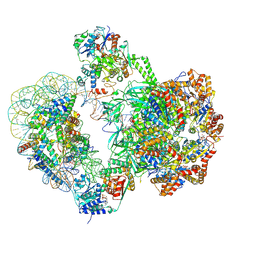 | | SWR1-nucleosome complex in configuration 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Nucleosome flipping drives kinetic proofreading and processivity by SWR1.
Nature, 636, 2024
|
|
8QKU
 
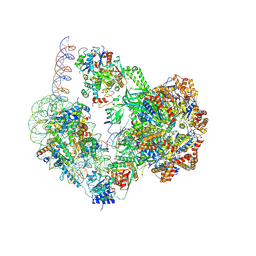 | | SWR1-nucleosome complex in configuration 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nucleosome flipping drives kinetic proofreading and processivity by SWR1.
Nature, 636, 2024
|
|
8QYV
 
 | | SWR1-hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
7EGP
 
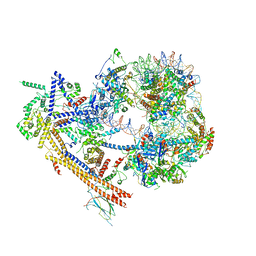 | | The structure of SWI/SNF-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Chen, Z.C, Chen, K.J, He, Z.Y, Ye, Y.P. | | Deposit date: | 2021-03-24 | | Release date: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity.
Cell Discov, 7, 2021
|
|
6KW3
 
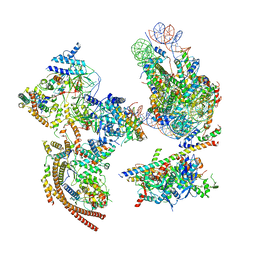 | | The ClassA RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
8B3I
 
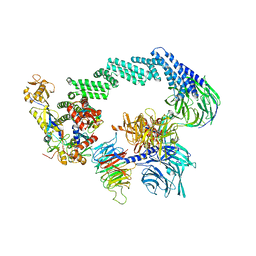 | | CRL4CSA-E2-Ub (state 2) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
5QS7
 
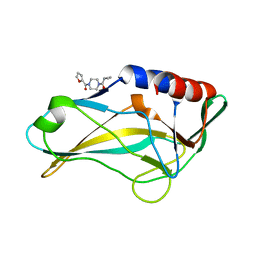 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z32327641 | | Descriptor: | T-box transcription factor T, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QSD
 
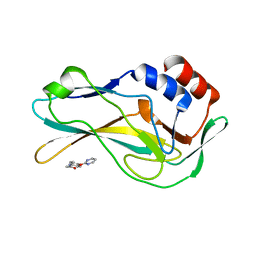 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z54571979 | | Descriptor: | N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QSK
 
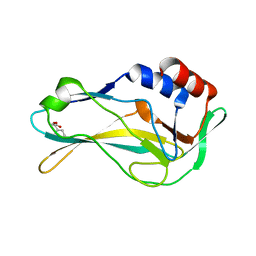 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z2856434906 | | Descriptor: | (benzyloxy)acetic acid, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QS9
 
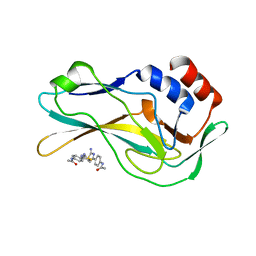 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z48847594 | | Descriptor: | N-[4-(2-amino-1,3-thiazol-4-yl)phenyl]acetamide, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5HYN
 
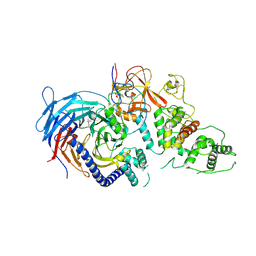 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with oncogenic histone H3K27M peptide | | Descriptor: | H3K27M, Histone-lysine N-methyltransferase EZH2, JARID2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Wilson, J.R, Gamblin, S.J. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Nat Commun, 7, 2016
|
|
8VML
 
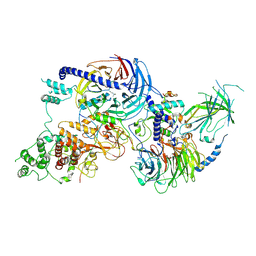 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VMI
 
 | | PRC2_AJ119-450 bound to H3K4me3 | | Descriptor: | EZH2, Histone H3.1, Histone H3.1t, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VNV
 
 | |
9FBW
 
 | | SWR1 lacking Swc5 subunit in complex with hexasome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2024-05-14 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
8OL1
 
 | |
6IRO
 
 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5K35
 
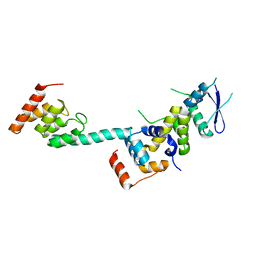 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
7EYQ
 
 | |
8JAQ
 
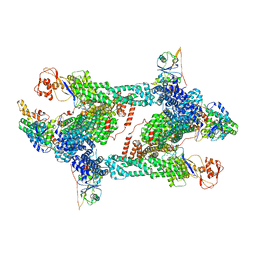 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MSJ
 
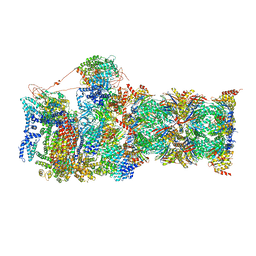 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
8JAS
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MSG
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
