4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
2ACV
 
 | | Crystal Structure of Medicago truncatula UGT71G1 | | Descriptor: | URIDINE-5'-DIPHOSPHATE, triterpene UDP-glucosyl transferase UGT71G1 | | Authors: | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
2ACW
 
 | | Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, triterpene UDP-glucosyl transferase UGT71G1 | | Authors: | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
2CVF
 
 | | Crystal structure of the RadB recombinase | | Descriptor: | DNA repair and recombination protein radB | | Authors: | Akiba, T, Ishii, N, Rashid, N, Morikawa, M, Imanaka, T, Harata, K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of RadB recombinase from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1: an implication for the formation of a near-7-fold helical assembly
Nucleic Acids Res., 33, 2005
|
|
2A0I
 
 | | F Factor TraI Relaxase Domain bound to F oriT Single-stranded DNA | | Descriptor: | F plasmid single-stranded oriT DNA, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Larkin, C, Datta, S, Harley, M.J, Anderson, B.J, Ebie, A, Hargreaves, V, Schildbach, J.F. | | Deposit date: | 2005-06-16 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the f factor relaxase.
Structure, 13, 2005
|
|
2AUN
 
 | |
3P0R
 
 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
4JG9
 
 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
4K2E
 
 | | HlyU from Vibrio cholerae N16961 | | Descriptor: | Transcriptional activator HlyU | | Authors: | Mukherjee, D, Datta, A.B, Chakrabarti, P. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of HlyU, the hemolysin gene transcription activator, from Vibrio cholerae N16961 and functional implications.
Biochim.Biophys.Acta, 1844, 2014
|
|
2YIX
 
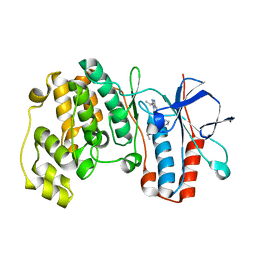 | | Triazolopyridine Inhibitors of p38 | | Descriptor: | 1-ethyl-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Han, s, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthawaite, R.A, Mahke, A, Marr, E, Mathias, J.P, Philip, J, Phillips, C, Smith, R.T, Stefaniak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
4KIN
 
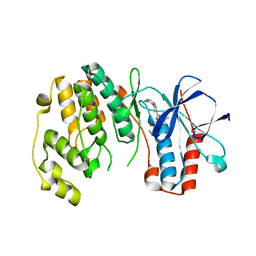 | |
4KIQ
 
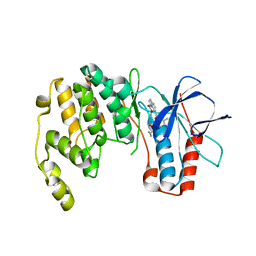 | |
2YIW
 
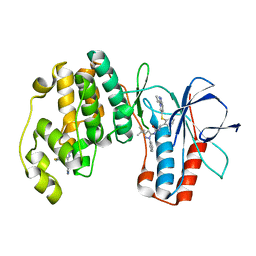 | | triazolopyridine inhibitors of p38 kinase | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthwaite, R.A, Mahnke, A, Mathais, J.P, Philip, J, Phillips, C, Smith, R.T, Stefamiak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
4KIP
 
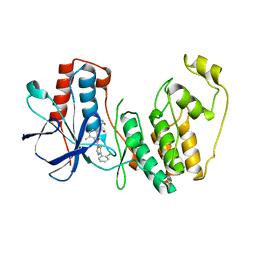 | |
4L7V
 
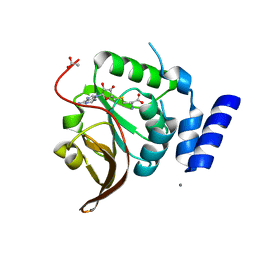 | | Crystal structure of Protein L-isoaspartyl-O-methyltransferase of Vibrio cholerae | | Descriptor: | ACETATE ION, CALCIUM ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Chatterjee, T, Mukherjee, D, Chakrabarti, P. | | Deposit date: | 2013-06-14 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure and activity of protein L-isoaspartyl-O-methyltransferase from Vibrio cholerae, and the effect of AdoHcy binding.
Arch.Biochem.Biophys., 583, 2015
|
|
4KWC
 
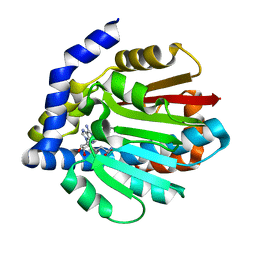 | |
3WIP
 
 | | Crystal structure of acetylcholine bound to Ls-AChBP | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholine-binding protein, ... | | Authors: | Olsen, J.A, Balle, T, Gajhede, M, Ahring, P.K, Kastrup, J.S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of the neurotransmitter acetylcholine by an acetylcholine binding protein reveals determinants of binding to nicotinic acetylcholine receptors
Plos One, 9, 2014
|
|
8RIU
 
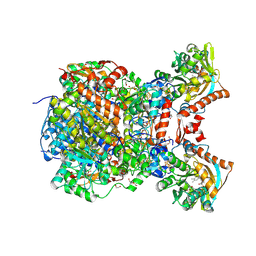 | |
8RJA
 
 | | Crystal structure of the F420-reducing formylmethanofuran dehydrogenase complex from the ethanotroph Candidatus Ethanoperedens thermophilum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-12-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Ethane-oxidising archaea couple CO 2 generation to F 420 reduction.
Nat Commun, 15, 2024
|
|
4EHI
 
 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
8RMG
 
 | |
8RMF
 
 | |
8RME
 
 | |
3ZH6
 
 | |
8RMC
 
 | |
