8F9M
 
 | |
9F0P
 
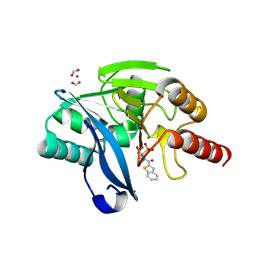 | | VIM-2 in complex with GKV61 (5c) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
9HWS
 
 | | Crystal structure of the Keap1 Kelch domain in complex with the small molecule UCAB#909 at 1.61 Angstrom resolution | | Descriptor: | 3-[7-[2-(cycloheptylamino)-2-oxidanylidene-ethoxy]naphthalen-2-yl]propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2025-01-06 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Novel High-affinity, Selective, and Anti-inflammatory Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9HWQ
 
 | | Crystal structure of the Keap1 Kelch domain in complex with the XChem fragment Z19735904 at 1.14 Angstrom resolution. | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2025-01-06 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment-Based Drug Discovery of Novel High-affinity, Selective, and Anti-inflammatory Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7TB0
 
 | | E. faecium MurAA in complex with fosfomycin and UNAG | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
8IOV
 
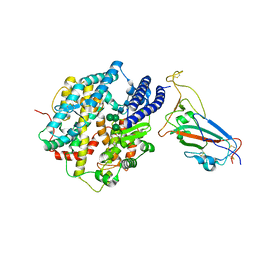 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
6MHR
 
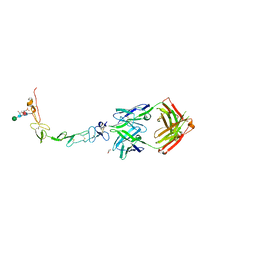 | | Structure of the human 4-1BB / Urelumab Fab complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
5X39
 
 | |
5X34
 
 | |
7TCV
 
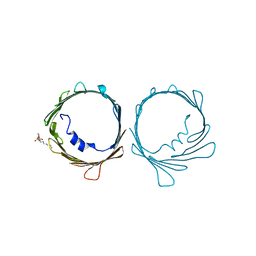 | | VDAC K12E mutant | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Khan, F, Abramson, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Dynamical control of the mitochondrial beta-barrel channel VDAC by electrostatic and mechanical coupling
To Be Published
|
|
5F0M
 
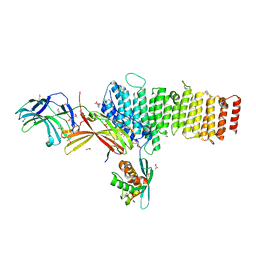 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
3KRZ
 
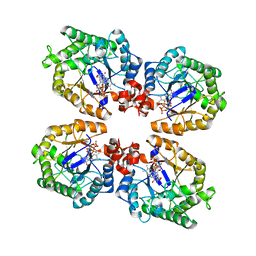 | | Crystal Structure of the Thermostable NADH4-bound old yellow enzyme from Thermoanaerobacter pseudethanolicus E39 | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-11-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
6D9M
 
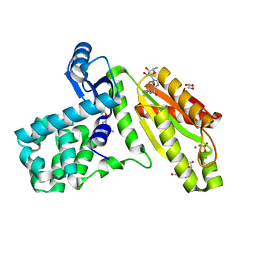 | | T4-Lysozyme fusion to Geobacter GGDEF | | Descriptor: | ACETATE ION, Fusion protein of Endolysin,Response receiver sensor diguanylate cyclase, GAF domain-containing, ... | | Authors: | Hallberg, Z, Doxzen, K, Kranzusch, P.J, Hammond, M. | | Deposit date: | 2018-04-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration.
Elife, 8, 2019
|
|
9IC2
 
 | | Structure of AMPK-alpha2-kinase domain bound to BAY-3827 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ahangar, M.S, Zeqiraj, E, Fraguas Bringas, C, Sakamoto, K. | | Deposit date: | 2025-02-14 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of AMPK-alpha2-kinase domain bound to BAY-3827
To Be Published
|
|
5F79
 
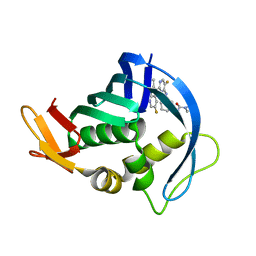 | | Influenza PB2 bound to an azaindole inhibitor | | Descriptor: | N-[(1R,3S)-3-({5-fluoro-2-[5-fluoro-2-(hydroxymethyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]pyrimidin-4-yl}amino)cyclohexyl]pyrrolidine-1-carboxamide, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 2-Substituted 7-Azaindole and 7-Azaindazole Analogues as Potential Antiviral Agents for the Treatment of Influenza.
ACS Med Chem Lett, 8, 2017
|
|
5F7G
 
 | |
5F27
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 2 at 1.68A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}-methyl-1-(4-piperidin-1-ylphenyl)methanamine | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
6P12
 
 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94131851 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
3GK0
 
 | |
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4QDK
 
 | |
5ZLA
 
 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
2MW1
 
 | |
