2W4X
 
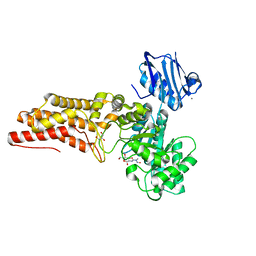 | | BtGH84 in complex with STZ | | Descriptor: | CALCIUM ION, GLYCEROL, O-GLCNACASE BT_4395, ... | | Authors: | He, Y, Bubb, A, Martinez-Fleites, C, Davies, G.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Insight Into the Mechanism of Streptozotocin Inhibition of O-Glcnacase.
Carbohydr.Res., 344, 2009
|
|
2W67
 
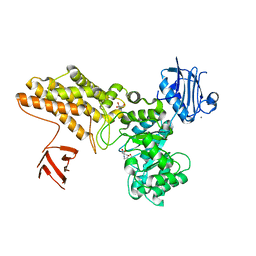 | | BtGH84 in complex with FMA34 | | Descriptor: | CALCIUM ION, GLYCEROL, N-[(3S,4R,5R,6R)-4,5,6-trihydroxyazepan-3-yl]acetamide, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2008-12-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Basis for Inhibition of Gh84 Glycoside Hydrolases by Substituted Azepanes: Conformational Flexibility Enables Probing of Substrate Distortion.
J.Am.Chem.Soc., 131, 2009
|
|
2VVN
 
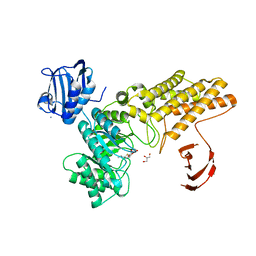 | | BtGH84 in complex with NH-Butylthiazoline | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, AMMONIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Potent Mechanism-Inspired O-Glcnacase Inhibitor that Blocks Phosphorylation of Tau in Vivo.
Nat.Chem.Biol., 4, 2008
|
|
2VVS
 
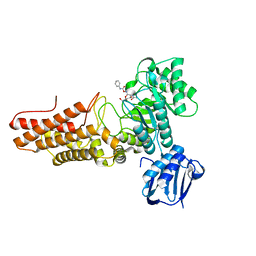 | | BtGH84 structure in complex with PUGNAc | | Descriptor: | O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GLCNACASE BT_4395 | | Authors: | Macauley, M.S, Bubb, A, Martinez-Fleites, C, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2008-06-11 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Elevation of Global O-Glcnac Levels in 3T3-L1 Adipocytes by Selective Inhibition of O-Glcnacase Does not Induce Insulin Resistance
J.Biol.Chem., 283, 2008
|
|
2WJZ
 
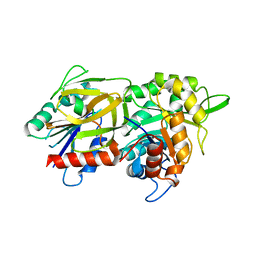 | | Crystal structure of (HisH) K181A Y138A mutant of imidazoleglycerolphosphate synthase (HisH HisF) which displays constitutive glutaminase activity | | Descriptor: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE HISF, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISH, PHOSPHATE ION | | Authors: | Vega, M.C, List, F, Razeto, A, Haeger, M.C, Babinger, K, Kuper, J, Sterner, R, Wilmanns, M. | | Deposit date: | 2009-06-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
6RH8
 
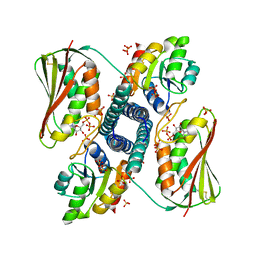 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RFV
 
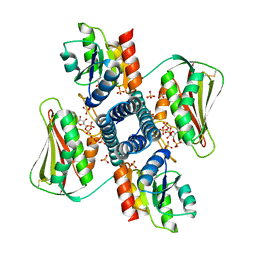 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
4FX7
 
 | |
4GTC
 
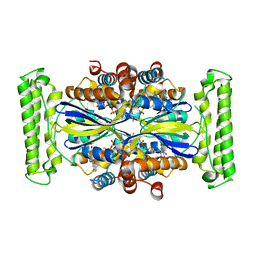 | | T. Maritima FDTS (E144R mutant) plus FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A, Prabhakar, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1W5F
 
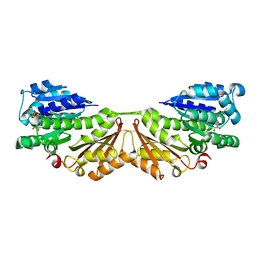 | | FtsZ, T7 mutated, domain swapped (T. maritima) | | Descriptor: | CELL DIVISION PROTEIN FTSZ, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
7NDZ
 
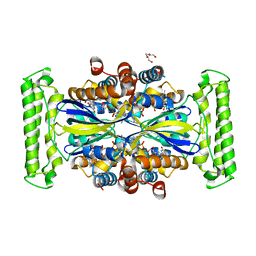 | | ThyX reconstituted with N5-carbinolamine flavin | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7NDW
 
 | | ThyX-FADH2 soaked with 20 mM Formaldehyde | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
8HF3
 
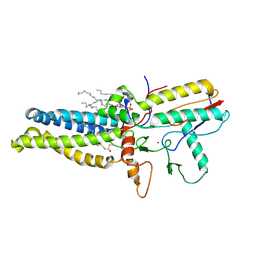 | | Cryo-EM structure of human ZDHHC9/GCP16 complex | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Golgin subfamily A member 7, PALMITIC ACID, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Liu, S, Yang, A. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4QCC
 
 | |
3CNN
 
 | | GTP-bound structure of TM YlqF | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3DCL
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
1GXK
 
 | |
1GXL
 
 | |
1GXJ
 
 | |
7KQ5
 
 | |
2W6R
 
 | | Crystal structure of an artificial (ba)8-barrel protein designed from identical half barrels | | Descriptor: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF | | Authors: | Hocker, B, Lochner, A, Seitz, T, Claren, J, Sterner, R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Crystal Structure of an Artificial (Betaalpha)(8)-Barrel Protein Designed from Identical Half-Barrels.
Biochemistry, 48, 2009
|
|
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | Descriptor: | COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5IOS
 
 | |
5IOT
 
 | |
