2PQU
 
 | |
2PY9
 
 | |
2LNG
 
 | |
2LNF
 
 | |
2LNE
 
 | |
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
2LMZ
 
 | |
2MH5
 
 | | Structure and NMR assignments of lantibiotic NAI-107 in DPC micelles | | Descriptor: | Lantibiotic 107891, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Munch, D, Muller, A, Schneider, T, Kohl, B, Wenzel, M, Bandow, J, Maffioli, S, Sosio, M, Donadio, S, Wimmer, R, Sahl, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The Lantibiotic NAI-107 Binds to Bactoprenol-bound Cell Wall Precursors and Impairs Membrane Functions.
J.Biol.Chem., 289, 2014
|
|
2M7R
 
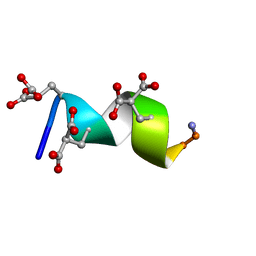 | | Nmda receptor antagonist, conantokin bk-b, nmr, 20 structure | | Descriptor: | CONANTOKIN BK-B | | Authors: | Curtice, K.J, Platt, R.J, Skalicky, J.J, Horvath, M.P, Twede, V.D. | | Deposit date: | 2013-04-29 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes.
Toxicon, 81C, 2014
|
|
2MX2
 
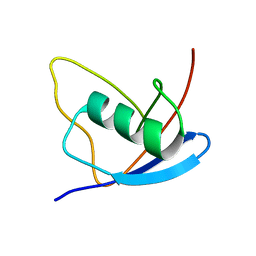 | | UBX-L domain of VCIP135 | | Descriptor: | Deubiquitinating protein VCIP135 | | Authors: | Iwazu, T, Murayama, S, Igarashi, R, Hrioaki, H, Shirakawa, M, Tochio, H. | | Deposit date: | 2014-12-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction mode of the UBX-L domain of VCIP135 determined by solution NMR spectroscopy
To be Published
|
|
2MX6
 
 | |
2MU0
 
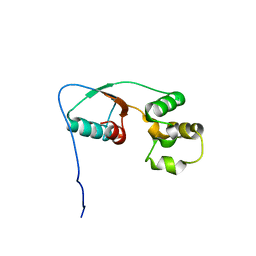 | |
2N9A
 
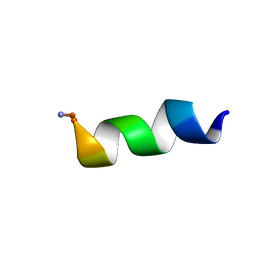 | |
2MZF
 
 | | Purotoxin-2 NMR structure in water | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Vassilevski, A, Oparin, P, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2OPM
 
 | | Human Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthetase (FPP synthetase) (FPS) (Farnesyl diphosphate synthetase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10)], MAGNESIUM ION, ... | | Authors: | Cao, R, Gao, Y.G, Robinson, H, Goddard, A. | | Deposit date: | 2007-01-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
2MPE
 
 | |
2N5Y
 
 | | Solution NMR structure of octyl-tridecaptin A1 in DPC micelles containing Gram-negative lipid II | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
2MZG
 
 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2MXZ
 
 | |
2L3X
 
 | |
2LK5
 
 | | Solution structure of the Zn(II) form of Desulforedoxin | | Descriptor: | Desulforedoxin, ZINC ION | | Authors: | Goodfellow, B.J, Tavares, P, Romao, M.J, Czaja, C, Rusnak, F, Legall, J, Moura, I, Moura, J.J.G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of desulforedoxin, a simple iron-sulfur protein - An NMR study of the zinc derivative
J.BIOL.INORG.CHEM., 1, 1996
|
|
2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
2MJ0
 
 | |
2MK3
 
 | | Solution NMR structure of gp41 ectodomain monomer on a DPC micelle | | Descriptor: | Transmembrane glycoprotein, chimeric construct | | Authors: | Roche, J, Louis, J.M, Grishaev, A, Ying, J, Bax, A. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
