6T5X
 
 | | Crystal structure of Salmonella typhimurium FabG in complex with NADPH at 1.5 A resolution | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Vella, P, Schnell, R, Schneider, G. | | 登録日 | 2019-10-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6T62
 
 | |
6T6P
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) at 1.57 A resolution | | 分子名称: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, PHOSPHATE ION | | 著者 | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | 登録日 | 2019-10-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6T7M
 
 | |
6T60
 
 | |
6T6N
 
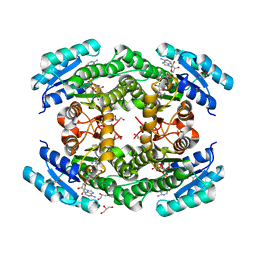 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | 著者 | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | 登録日 | 2019-10-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6T77
 
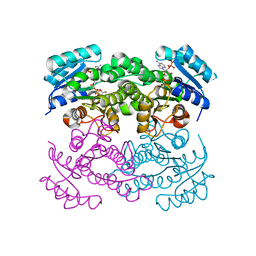 | | Crystal structure of Klebsiella pneumoniae FabG(NADPH-dependent) NADP-complex at 1.75 A resolution | | 分子名称: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | 登録日 | 2019-10-21 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
3FUB
 
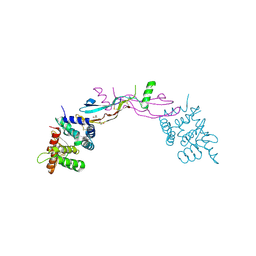 | | Crystal structure of GDNF-GFRalpha1 complex | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Parkash, V, Goldman, A. | | 登録日 | 2009-01-14 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Comparison of GFL-GFRalpha complexes: further evidence relating GFL bend angle to RET signalling
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7AXZ
 
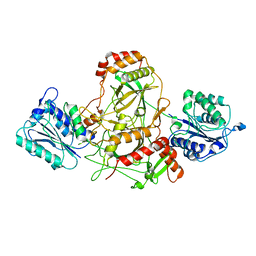 | | Ku70/80 complex apo form | | 分子名称: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | 著者 | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | 登録日 | 2020-11-10 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
8EJN
 
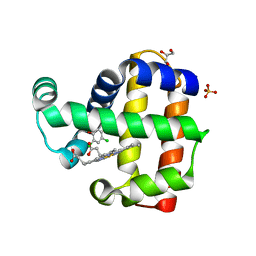 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | 分子名称: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | 著者 | Aktar, M.S, de Serrano, V.S, Franzen, S. | | 登録日 | 2022-09-17 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.481 Å) | | 主引用文献 | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | 著者 | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | 登録日 | 2020-12-09 | | 公開日 | 2021-04-07 | | 最終更新日 | 2021-08-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
6TV2
 
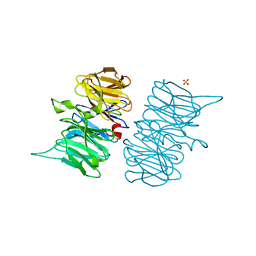 | | Heme d1 biosynthesis associated Protein NirF | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | 著者 | Kluenemann, T, Layer, G, Blankenfeldt, W. | | 登録日 | 2020-01-08 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.561 Å) | | 主引用文献 | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
6TYR
 
 | |
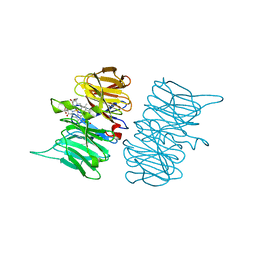
6TV9
 
 | |
6UBQ
 
 | |
6UCW
 
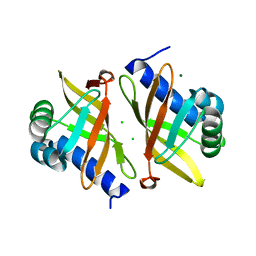 | | Multi-conformer model of Apo Ketosteroid Isomerase from Pseudomonas Putida (pKSI) at 250 K | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Steroid Delta-isomerase | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-17 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TZD
 
 | |
6EZW
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group C2 | | 分子名称: | VHH antibody BCD090-M2 | | 著者 | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | 登録日 | 2017-11-16 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.59842849 Å) | | 主引用文献 | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
6U4I
 
 | |
6U1Z
 
 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | 著者 | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | 登録日 | 2017-11-07 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7BR9
 
 | | Crystal structure of mus musculus IRG1 | | 分子名称: | Cis-aconitate decarboxylase | | 著者 | Park, H.H, Chun, H.L. | | 登録日 | 2020-03-27 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The crystal structure of mouse IRG1 suggests that cis-aconitate decarboxylase has an open and closed conformation.
Plos One, 15, 2020
|
|
6EVG
 
 | |
6UCN
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to Equilenin at 250 K | | 分子名称: | CHLORIDE ION, EQUILENIN, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UCY
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to 4-Androstenedione at 250 K | | 分子名称: | 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
