3PIG
 
 | | beta-fructofuranosidase from Bifidobacterium longum | | Descriptor: | Beta-fructofuranosidase, CHLORIDE ION | | Authors: | Bujacz, A, Bujacz, G, Redzynia, I, Krzepkowska-Jedrzejczak, M, Bielecki, S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the apo form of beta-fructofuranosidase from Bifidobacterium longum and its complex with fructose
Febs J., 278, 2011
|
|
3RWK
 
 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | ACETATE ION, Inulinase, SODIUM ION, ... | | Authors: | Michaux, C, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Housen, I, Wouters, J. | | Deposit date: | 2011-05-09 | | Release date: | 2012-07-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
3SC7
 
 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Inulinase, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Housen, I, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Wouters, J, Michaux, C. | | Deposit date: | 2011-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
3U14
 
 | | Structure of D50A-fructofuranosidase from Schwanniomyces occidentalis complexed with inulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
4EQV
 
 | |
3U75
 
 | | Structure of E230A-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosylnystose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
3UGG
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 1-kestose | | Descriptor: | GLYCEROL, SULFATE ION, Sucrose:(Sucrose/fructan) 6-fructosyltransferase, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UGH
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
6NU7
 
 | |
3UGF
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
6NU8
 
 | |
6NUN
 
 | |
6NUM
 
 | |
4FFH
 
 | |
4FFG
 
 | | Crystal Structure of Levan Fructotransferase from Arthrobacter ureafaciens in complex with DFA-IV | | Descriptor: | (1R,4R,5S,6S,7R,10R,11S,12S)-1,7-bis(hydroxymethyl)-2,8,13,14-tetraoxatricyclo[8.2.1.1~4,7~]tetradecane-5,6,11,12-tetrol, Levan fructotransferase, beta-D-fructofuranose-(2-6)-beta-D-fructofuranose | | Authors: | Park, J, Rhee, S. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional basis for substrate specificity and catalysis of levan fructotransferase.
J.Biol.Chem., 287, 2012
|
|
4FFF
 
 | |
4FFI
 
 | |
3LF7
 
 | |
3LDK
 
 | | Crystal Structure of A. japonicus CB05 | | Descriptor: | Fructosyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chuankhayan, P, Chen, C.J, Chaing, C.M, Hsieh, C.Y, Chen, C.D, Hsieh, Y.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete sbusites in the active site for catalysis
To be Published
|
|
3LIH
 
 | |
8AA0
 
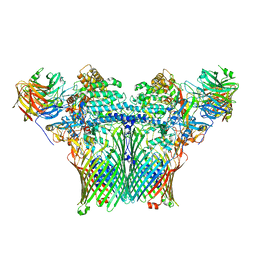 | | Levan utilisation machinery (utilisome) with levan fructo-oligosaccharides DP 8-12 | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
3LIG
 
 | |
8AA2
 
 | | Inactive levan utilisation machinery (utilisome) in the presence of levan fructo-oligosaccharides DP 15-25 | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8A9Y
 
 | | Substrate-free levan utilisation machinery (utilisome) | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
3LEM
 
 | |
