1XAX
 
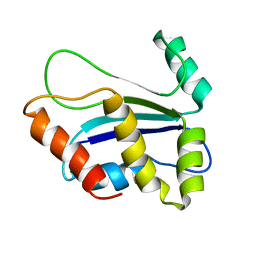 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
1XKX
 
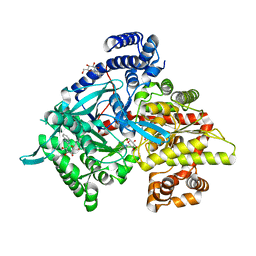 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | 2-(BETA-D-GLUCOPYRANOSYL)-5-METHYL-1-BENZIMIDAZOLE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1XL0
 
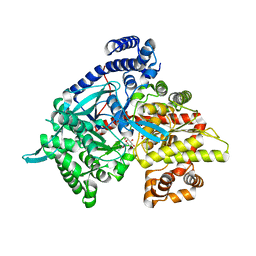 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3,4-oxadiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1XL1
 
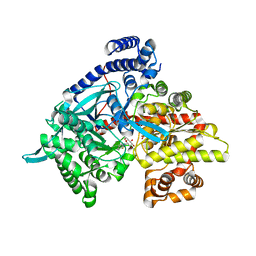 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3-benzothiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
5N0L
 
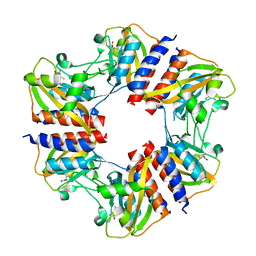 | | The structure of the cofactor binding GAF domain of the nutrient sensor CodY from Clostridium difficile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Sonenshein, A.L. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.
Plos One, 14, 2019
|
|
1AAL
 
 | |
2V91
 
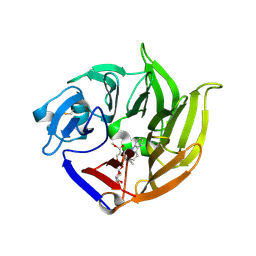 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH STRICTOSIDINE | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE SYNTHASE | | Authors: | Loris, E.A, Panjikar, S, Ruppert, M, Barleben, L, Unger, M, Stoeckigt, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure Based Engineering of Strictosidine Synthase: Auxiliary for Alkaloid Libraries
Chem.Biol., 14, 2007
|
|
3C2H
 
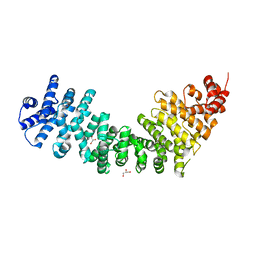 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3C2G
 
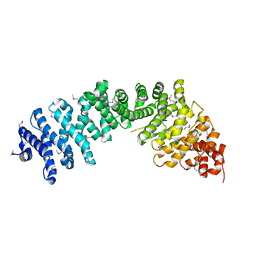 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
1PTD
 
 | |
1PTG
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH MYO-INOSITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Ryan, M, Bullock, T.L, Griffith, O.H. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
EMBO J., 14, 1995
|
|
1ZDM
 
 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
1ZCP
 
 | |
2A8P
 
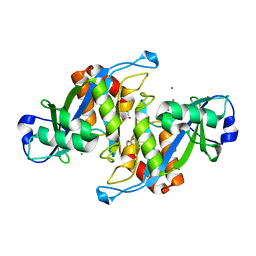 | | 2.7 Angstrom Crystal Structure of the Complex Between the Nuclear SnoRNA Decapping Nudix Hydrolase X29 and Manganese | | Descriptor: | MANGANESE (II) ION, U8 snoRNA-binding protein X29 | | Authors: | Scarsdale, J.N, Peculis, B.A, Wright, H.T. | | Deposit date: | 2005-07-08 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes
Structure, 14, 2006
|
|
2A38
 
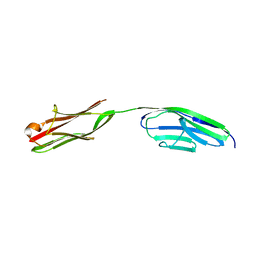 | | Crystal structure of the N-Terminus of titin | | Descriptor: | CADMIUM ION, Titin | | Authors: | Marino, M, Muhle-Goll, C, Svergun, D, Demirel, M, Mayans, O. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin
Structure, 14, 2006
|
|
2ALA
 
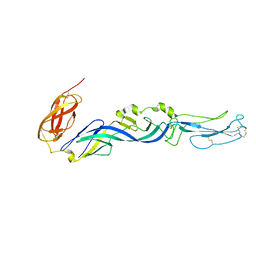 | | Crystal structure of the Semliki Forest Virus envelope protein E1 in its monomeric conformation. | | Descriptor: | Structural polyprotein (P130) | | Authors: | Roussel, A, Lescar, J, Vaney, M.C, Wengler, G, Wengler, G, Rey, F.A. | | Deposit date: | 2005-08-05 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus.
Structure, 14, 2006
|
|
2B3X
 
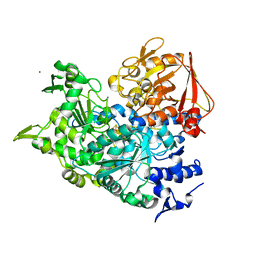 | | Structure of an orthorhombic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, Iron-responsive element binding protein 1, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
1ZW6
 
 | | Crystal Structure of the GTP-bound form of RasQ61G | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ford, B, Hornak, V, Kleinman, H, Nassar, N. | | Deposit date: | 2005-06-03 | | Release date: | 2006-03-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a transient intermediate for GTP hydrolysis by ras.
Structure, 14, 2006
|
|
2BEV
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-2-METHYL-BUTYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2ABK
 
 | |
2A8T
 
 | | 2.1 Angstrom Crystal Structure of the Complex Between the Nuclear U8 snoRNA Decapping Nudix Hydrolase X29, Manganese and m7G-PPP-A | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE, MANGANESE (II) ION, ... | | Authors: | Scarsdale, J.N, Peculis, B.A, Wright, H.T. | | Deposit date: | 2005-07-08 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes
Structure, 14, 2006
|
|
1RXP
 
 | |
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
1ZVQ
 
 | |
1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
