1AD3
 
 | |
1AJL
 
 | |
1AF3
 
 | | RAT BCL-XL AN APOPTOSIS INHIBITORY PROTEIN | | Descriptor: | APOPTOSIS REGULATOR BCL-X | | Authors: | Aritomi, M, Kunishima, N, Inohara, N, Ishibashi, Y, Ohta, S, Morikawa, K. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family.
J.Biol.Chem., 272, 1997
|
|
1AC9
 
 | | SOLUTION STRUCTURE OF A DNA DECAMER CONTAINING THE ANTIVIRAL DRUG GANCICLOVIR: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS, AND FULL RELAXATION REFINEMENT, 6 STRUCTURES | | Descriptor: | DNA | | Authors: | Foti, M, Marshalko, S, Schurter, E, Kumar, S, Beardsley, G.P, Schweitzer, B.I. | | Deposit date: | 1997-02-17 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer containing the antiviral drug ganciclovir: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 36, 1997
|
|
1DJX
 
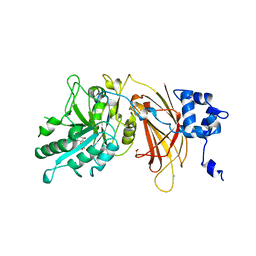 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJG
 
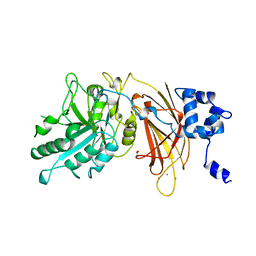 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH LANTHANUM | | Descriptor: | ACETATE ION, LANTHANUM (III) ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1AIS
 
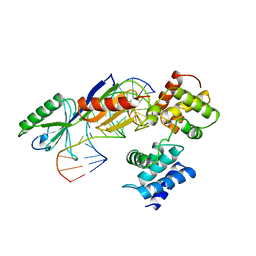 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*AP*CP*TP*TP*TP*(5IU)P*(5IU)P*AP*AP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*AP*AP*GP*TP*T )-3'), PROTEIN (TATA-BINDING PROTEIN), ... | | Authors: | Kosa, P.F, Ghosh, G, Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1997-04-24 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ACZ
 
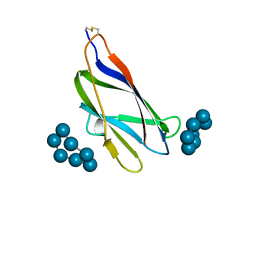 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, 5 STRUCTURES | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLUCOAMYLASE | | Authors: | Sorimachi, K, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1997-02-10 | | Release date: | 1997-07-07 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to beta-cyclodextrin.
Structure, 5, 1997
|
|
1AJ3
 
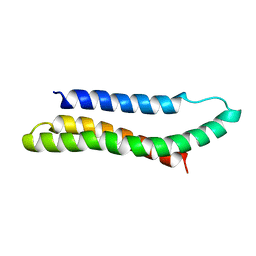 | | SOLUTION STRUCTURE OF THE SPECTRIN REPEAT, NMR, 20 STRUCTURES | | Descriptor: | ALPHA SPECTRIN | | Authors: | Pascual, J, Pfuhl, M, Walther, D, Saraste, M, Nilges, M. | | Deposit date: | 1997-05-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the spectrin repeat: a left-handed antiparallel triple-helical coiled-coil.
J.Mol.Biol., 273, 1997
|
|
1WKD
 
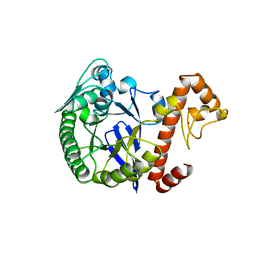 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKE
 
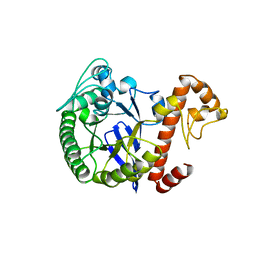 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1MKX
 
 | |
1WKF
 
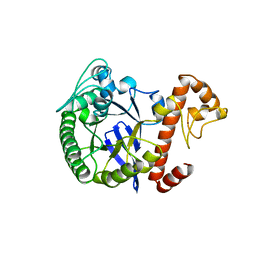 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1AIU
 
 | | HUMAN THIOREDOXIN (D60N MUTANT, REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Andersen, J.F, Gasdaska, J.R, Sanders, D.A.R, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-04-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
1AG3
 
 | |
1ABV
 
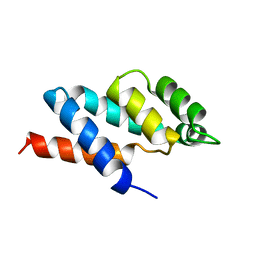 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | Authors: | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | Deposit date: | 1997-01-29 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
1AJF
 
 | |
1MIS
 
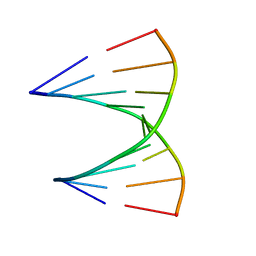 | |
2NEF
 
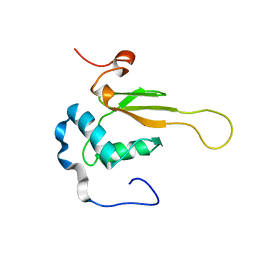 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
1TLS
 
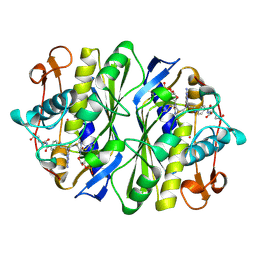 | | THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH FDUMP AND METHYLENETETRAHYDROFOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Hyatt, D.C, Maley, F, Montfort, W.R. | | Deposit date: | 1996-12-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate.
Biochemistry, 36, 1997
|
|
2REL
 
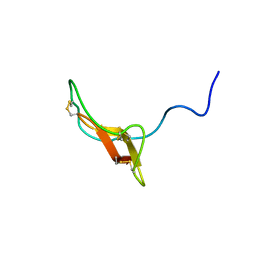 | | SOLUTION STRUCTURE OF R-ELAFIN, A SPECIFIC INHIBITOR OF ELASTASE, NMR, 11 STRUCTURES | | Descriptor: | R-ELAFIN | | Authors: | Francart, C, Dauchez, M, Alix, A.J.P, Lippens, G. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of R-elafin, a specific inhibitor of elastase.
J.Mol.Biol., 268, 1997
|
|
1EXN
 
 | | T5 5'-EXONUCLEASE | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Sayers, J.R, Stier, G, Suck, D. | | Deposit date: | 1997-01-17 | | Release date: | 1997-07-07 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A helical arch allowing single-stranded DNA to thread through T5 5'-exonuclease.
Nature, 382, 1996
|
|
1GAB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1996-12-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1DAP
 
 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ | | Descriptor: | ACETATE ION, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Reddy, S.G, Blanchard, J.S. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of meso-diaminopimelic acid dehydrogenase from Corynebacterium glutamicum.
Biochemistry, 35, 1996
|
|
3DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
