4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
4NOH
 
 | | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Lipoprotein, putative | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4NU7
 
 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | Descriptor: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NV4
 
 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
4JNN
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine | | Descriptor: | BENZAMIDINE, BETA-MERCAPTOETHANOL, Transcriptional regulator | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-15 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine
To be Published
|
|
4NLM
 
 | | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo1340 protein | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e
To be Published
|
|
4NEL
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine | | Descriptor: | N,N-dimethylmethanamine, Transcriptional regulator | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine
To be Published
|
|
4ISC
 
 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4IUO
 
 | | 1.8 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) K170M Mutant in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4NU9
 
 | | 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Stam, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O0N
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4O4A
 
 | | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Lipoprotein, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2HJQ
 
 | | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449 | | Descriptor: | Hypothetical protein yqbF | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449
TO BE PUBLISHED
|
|
4OVD
 
 | | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469 | | Descriptor: | CALCIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Babnigg, G, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469
To be Published
|
|
4JB7
 
 | | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Accessory colonization factor AcfC, D-MALATE, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Dubrovska, I, Winsor, J, Minasov, G, Shuvalova, L, Filippova, E.V, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid
To be Published
|
|
4JBF
 
 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
2GBS
 
 | | NMR structure of Rpa0253 from Rhodopseudomonas palustris. Northeast structural genomics consortium target RpR3 | | Descriptor: | Hypothetical protein Rpa0253 | | Authors: | Ramelot, T.A, Cort, J.R, Conover, K, Chen, Y, Ma, L.C, Ciano, M, Xiao, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-11 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
4JG2
 
 | | Structure of phage-related protein from Bacillus cereus ATCC 10987 | | Descriptor: | Phage-related protein | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of phage-related protein from Bacillus cereus ATCC 10987
To be Published
|
|
4JJX
 
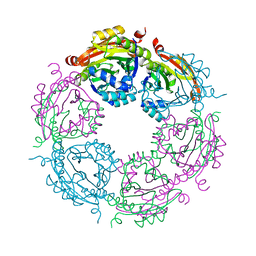 | | Dodecameric structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae O1 biovar eltor | | Descriptor: | Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4JJP
 
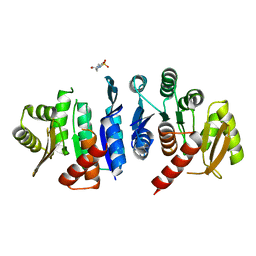 | | 2.06 Angstrom resolution crystal structure of phosphomethylpyrimidine kinase (thiD)from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phosphomethylpyrimidine kinase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | 2.06 Angstrom resolution crystal structure of phosphomethylpyrimidine kinase (thiD)from Clostridium difficile 630
To be Published
|
|
4JLY
 
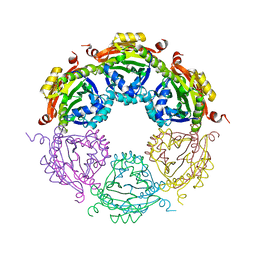 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4K15
 
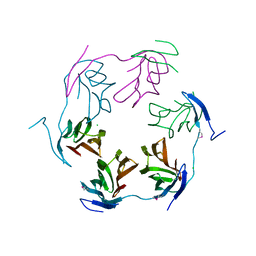 | | 2.75 Angstrom Crystal Structure of Hypothetical Protein lmo2686 from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, Lmo2686 protein | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Crystal Structure of Hypothetical Protein lmo2686 from Listeria monocytogenes EGD-e
TO BE PUBLISHED
|
|
2FN1
 
 | | Crystal structures of Yersinia enterocolitica salicylate synthase (Irp9) in complex with the reaction products salicylate and pyruvate | | Descriptor: | 2-HYDROXYBENZOIC ACID, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kerbarh, O, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2006-01-10 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Yersinia enterocolitica Salicylate Synthase and its Complex with the Reaction Products Salicylate and Pyruvate.
J.Mol.Biol., 357, 2006
|
|
