4HG9
 
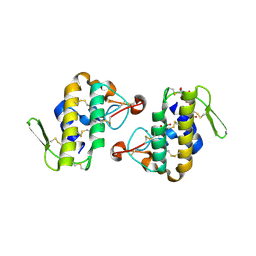 | | Crystal structure of AhV_bPA, a basic PLA2 from Agkistrodon halys pallas venom | | Descriptor: | Basic phospholipase A2 B, CALCIUM ION, CITRIC ACID, ... | | Authors: | Zeng, F, Niu, L, Li, X, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of AhV_bPA, a basic PLA2 from Agkistrodon halys pallas venom
TO BE PUBLISHED
|
|
1OK4
 
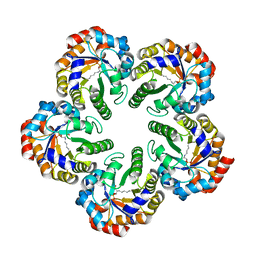 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK6
 
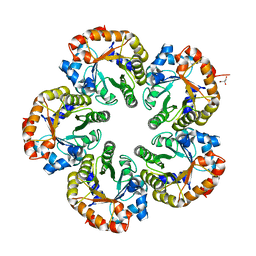 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OJ7
 
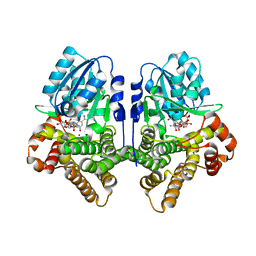 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
9C61
 
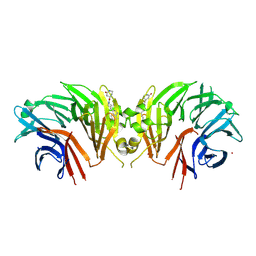 | | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26 | | Descriptor: | (4R)-4-[4-(5-fluoro-1H-indol-3-yl)piperidine-1-carbonyl]piperidin-2-one, Non-specific serine/threonine protein kinase, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Kutera, M, Ilyassov, O, Seitova, A, Loppnau, P, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-06-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26
To be published
|
|
3AMS
 
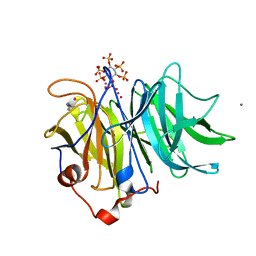 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
2X2T
 
 | | CRYSTAL STRUCTURE OF SCLEROTINIA SCLEROTIORUM AGGLUTININ SSA in complex with Gal-beta1,3-Galnac | | Descriptor: | AGGLUTININ, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
2JEA
 
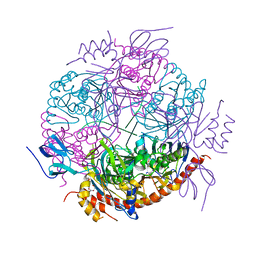 | |
2JEB
 
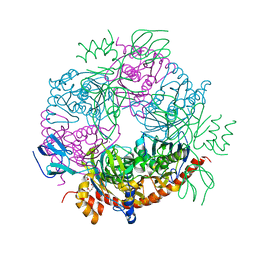 | |
4H0S
 
 | |
2ERS
 
 | |
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2X2S
 
 | | Crystal structure of Sclerotinia sclerotiorum agglutinin SSA | | Descriptor: | AGGLUTININ, GLYCEROL | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
4E7N
 
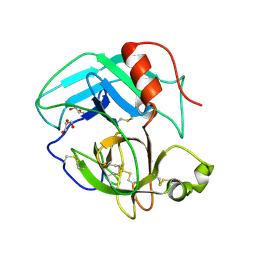 | | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Snake-venom Thrombin-like Enzyme | | Authors: | Zeng, F, Li, X, Teng, M, Niu, L. | | Deposit date: | 2012-03-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys
to be published
|
|
2JC6
 
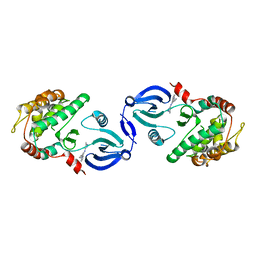 | | Crystal structure of human calmodulin-dependent protein kinase 1D | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE 1D, N-(5-METHYL-1H-PYRAZOL-3-YL)-2-PHENYLQUINAZOLIN-4-AMINE | | Authors: | Debreczeni, J.E, Rellos, P, Fedorov, O, Niesen, F.H, Bhatia, C, Shrestha, L, Salah, E, Smee, C, Colebrook, S, Berridge, G, Gileadi, O, Bunkoczi, G, Ugochukwu, E, Pike, A.C.W, von Delft, F, Knapp, S, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase 1D
To be Published
|
|
2JE6
 
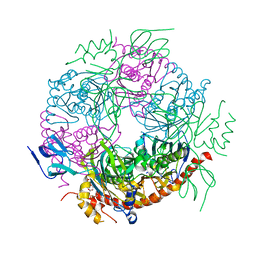 | | Structure of a 9-subunit archaeal exosome | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2007-01-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Channelling by the Archaeal Exosome.
Embo Rep., 8, 2007
|
|
2JAM
 
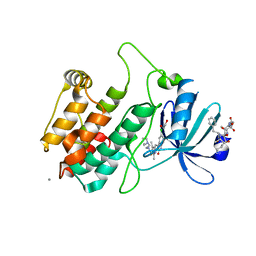 | | Crystal structure of human calmodulin-dependent protein kinase I G | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Keates, T, Niesen, F.H, Salah, E, Shrestha, L, Smee, C, Sobott, F, Pike, A.C.W, Bunkoczi, G, von Delft, F, Turnbull, A, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase I G
To be Published
|
|
5AKB
 
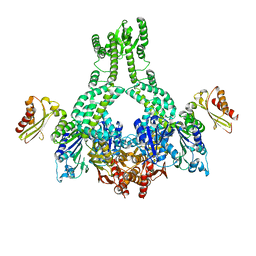 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKC
 
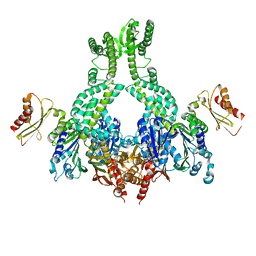 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKD
 
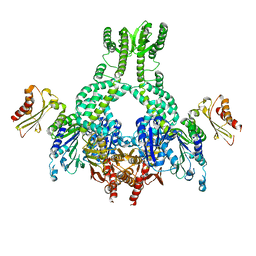 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
8HQF
 
 | |
8HVK
 
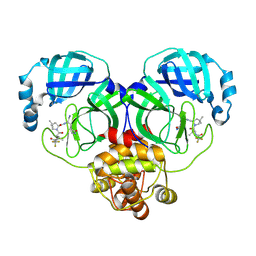 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
7WPQ
 
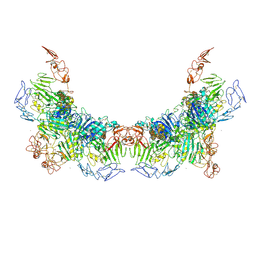 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 3.27 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.267 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WQT
 
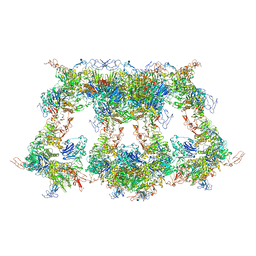 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (VWF tube) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
