9S2V
 
 | | NSP14 IN COMPLEX WITH LIGAND TDI-014925-CL-2 (compound 58) | | Descriptor: | CHLORIDE ION, Guanine-N7 methyltransferase nsp14, IMIDAZOLE, ... | | Authors: | Steinbacher, S, Huggins, D.J. | | Deposit date: | 2025-07-22 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery, Optimization, and Evaluation of Non-Nucleoside SARS-CoV-2 NSP14 Inhibitors.
J.Med.Chem., 68, 2025
|
|
9QCZ
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K263A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Active Site of Class 3 L-Asparaginase by Mutagenesis: Mutations of the Ser-Lys Tandems of ReAV.
Biomolecules, 15, 2025
|
|
7QRU
 
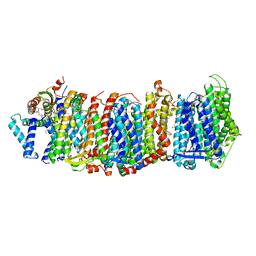 | | Structure of Bacillus pseudofirmus Mrp antiporter complex, monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Na(+)/H(+) antiporter subunit B, Na(+)/H(+) antiporter subunit C, ... | | Authors: | Lee, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations.
Nat Commun, 13, 2022
|
|
4R3R
 
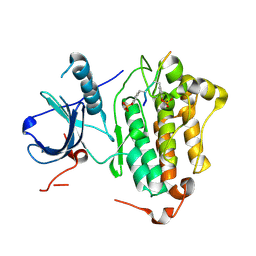 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7OKM
 
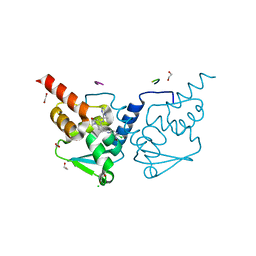 | | Crystal structure of human BCL6 BTB domain in complex with compound 13g | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-2-oxidanylidene-4-(2-pyrimidin-2-ylpropan-2-ylamino)quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
9N7A
 
 | |
7OI1
 
 | |
9S3A
 
 | | TaGST-10 in complex with deoxynivalenol-13-glutathione | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Deoxynivalenol-13-cysteine, ... | | Authors: | Michlmayr, H, Papageorgiou, A.C. | | Deposit date: | 2025-07-24 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Detoxification of deoxynivalenol by pathogen-inducible tau-class glutathione transferases from wheat.
J.Biol.Chem., 301, 2025
|
|
9QFD
 
 | | Cryo-EM structure of the fully cofilin-1-decorated actin filament (cofilactin) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-11 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
9O42
 
 | | Crystal structure of the L411A mutant of pregnane X receptor ligand binding domain in complex with SJPYT-328 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, Pregnane X receptor ligand binding domain tethered to steroid receptor coactivator-1 peptide | | Authors: | Huber, A.D, Garcia-Maldonado, E, Miller, D.J, Chen, T. | | Deposit date: | 2025-04-08 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Subtle changes in ligand-receptor interactions dramatically alter transcriptional outcomes of pregnane X receptor modulators.
Structure, 2025
|
|
7OKG
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8e | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(1-methylpyrazol-4-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
9NRA
 
 | |
8KEQ
 
 | |
9QWJ
 
 | | Crystal structure of S2c TCR in complex with CD1c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karuppiah, V. | | Deposit date: | 2025-04-14 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c + cells.
Front Immunol, 16, 2025
|
|
6FZO
 
 | | SMURFP-Y56F mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Janowski, R, Fuenzalida-Wernera, J.P, Mishra, K, Vetschera, P, Weidenfeld, I, Richter, K, Niessing, D, Ntziachristos, V, Stiel, A.C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation.
J. Struct. Biol., 204, 2018
|
|
5IX8
 
 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
4R2N
 
 | | Crystal structure of Rv3772 in complex with its substrate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
9O0J
 
 | | HtaA CR1 domain from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ford, J, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2025-04-02 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of heme scavenging by the ChtA and HtaA hemophores in Corynebacterium diphtheriae.
J.Biol.Chem., 301, 2025
|
|
9OML
 
 | |
9R4U
 
 | | Crystal structure of CtGH76 from Chaetomium thermophilum in complex with isomaltose | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein, ... | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9S71
 
 | |
9U7F
 
 | | structure of human KCNQ1-KCNE1-CaM complex | | Descriptor: | CALCIUM ION, Calmodulin-1, POTASSIUM ION, ... | | Authors: | Hou, P.P, Zhang, J, Wan, S.Y, Cheng, X.Y, Zhong, L, Hu, B. | | Deposit date: | 2025-03-24 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Secondary structure transitions and dual PIP2 binding define cardiac KCNQ1-KCNE1 channel gating.
Cell Res., 35, 2025
|
|
9QEY
 
 | | Cryo-EM structure of the actin filament bound by a single Coronin-1B molecule. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-11 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
9NCW
 
 | | Crystal Structure of WDR5 in complex with Triazole-Based Inhibitors | | Descriptor: | 1-{[(6P)-3-[(3,5-dimethoxyphenyl)methyl]-6-(4-fluoro-2-methylphenyl)-4,5-dihydro-3H-naphtho[1,2-d][1,2,3]triazol-8-yl]methyl}-3-methyl-1,3-dihydro-2H-imidazol-2-imine, WD repeat-containing protein 5 | | Authors: | Goins, C.M, Stauffer, S.R. | | Deposit date: | 2025-02-17 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Development and Characterization of Triazole-Based WDR5 Inhibitors for the Treatment of Glioblastoma
Biorxiv, 2025
|
|
9PBB
 
 | | 293K human S-adenosylmethionine decarboxylase | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Patel, J.R, Bonzon, T.J, Bahkt, T, Fagbohun, O.O, Clinger, J.A. | | Deposit date: | 2025-06-26 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions.
Biomolecules, 15, 2025
|
|
