1UJR
 
 | | Solution structure of WWE domain in BAB28015 | | Descriptor: | hypothetical protein AK012080 | | Authors: | He, F, Muto, Y, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-11 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in BAB28015
To be Published
|
|
1UJY
 
 | | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | He, F, Muto, Y, Uda, H, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-02-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6
To be Published
|
|
1UJU
 
 | |
1UJT
 
 | |
1UJX
 
 | |
1P5D
 
 | |
1P68
 
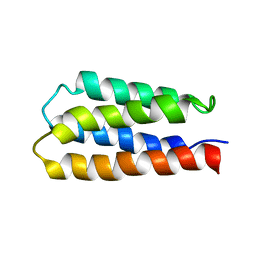 | | Solution structure of S-824, a de novo designed four helix bundle | | Descriptor: | De novo designed protein S-824 | | Authors: | Wei, Y, Kim, S, Fela, D, Baum, J, Hecht, M.H. | | Deposit date: | 2003-04-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a de novo protein from a designed combinatorial library.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1OXN
 
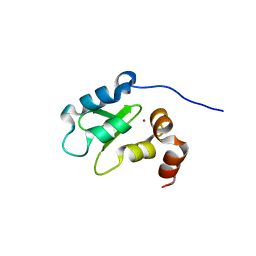 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEAVPWKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
2ANT
 
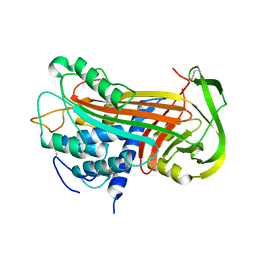 | | THE 2.6 A STRUCTURE OF ANTITHROMBIN INDICATES A CONFORMATIONAL CHANGE AT THE HEPARIN BINDING SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose, ANTITHROMBIN | | Authors: | Skinner, R, Abrahams, J.-P, Whisstock, J.C, Lesk, A.M, Carrell, R.W, Wardell, M.R. | | Deposit date: | 1997-01-28 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site.
J.Mol.Biol., 266, 1997
|
|
2AR0
 
 | |
2AWP
 
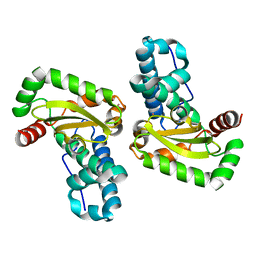 | | Crystal structure of Plasmodium knowlesi structure of Iron Super-Oxide Dismutase | | Descriptor: | CHLORIDE ION, Iron Super-Oxide Dismutase, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Zhao, Y, Lew, J, Alam, Z, Melone, M, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1ORQ
 
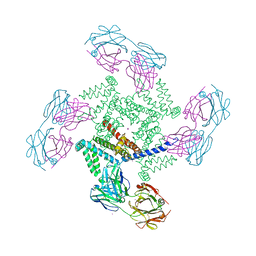 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1UYC
 
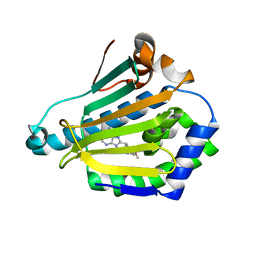 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
2AZ1
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum | | Descriptor: | CALCIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
2B76
 
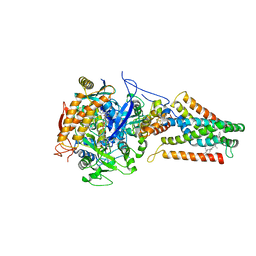 | | E. coli Quinol fumarate reductase FrdA E49Q mutation | | Descriptor: | CITRATE ANION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Maklashina, E, Iverson, T.M, Sher, Y, Kotlyar, V, Mirza, O, Andrell, J, Hudson, J.M, Armstrong, F.A, Cecchini, G. | | Deposit date: | 2005-10-03 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
J.Biol.Chem., 281, 2006
|
|
1OXS
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
2B8J
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with adenosine and phosphate at 2 A resolution | | Descriptor: | ADENOSINE, GOLD 3+ ION, GOLD ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Thaller, M.C, Rossolini, G.M, Mangani, S. | | Deposit date: | 2005-10-07 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | A structure-based proposal for the catalytic mechanism of the bacterial acid phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases
J.Mol.Biol., 355, 2006
|
|
2B9E
 
 | | Human NSUN5 protein | | Descriptor: | NOL1/NOP2/Sun domain family, member 5 isoform 2, S-ADENOSYLMETHIONINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Human NSUN5 protein in complex with
S-adenosyl-L-methionine
To be Published
|
|
2B9U
 
 | | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii | | Descriptor: | hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Rajakannan, V, Kondo, K, Mizushima, T, Suzuki, A, Yamane, T. | | Deposit date: | 2005-10-13 | | Release date: | 2006-10-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii
TO BE PUBLISHED
|
|
1MS9
 
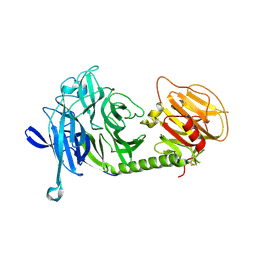 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme of Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
2ANA
 
 | |
1RWZ
 
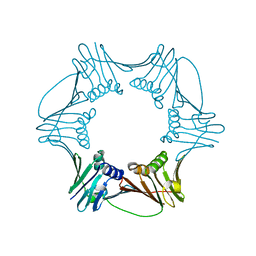 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
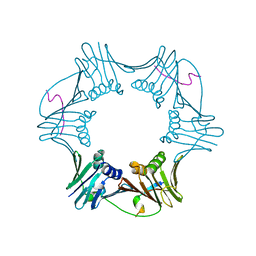 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
2CHO
 
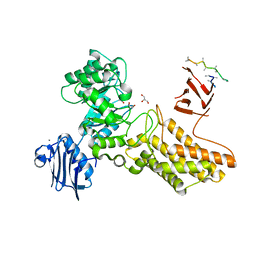 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
1N78
 
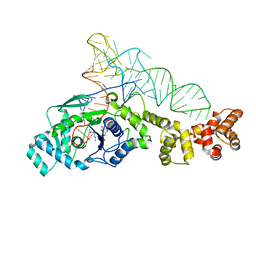 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and glutamol-AMP. | | Descriptor: | GLUTAMOL-AMP, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
