8U06
 
 | | Imine reductase RedE bound with NADP+ and arcyriaflavin A (primary site) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arcyriaflavin A, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An imine reductase that captures reactive intermediates in the biosynthesis of the indolocarbazole reductasporine.
J.Biol.Chem., 300, 2024
|
|
7PZL
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZM
 
 | | HBc-P5T in complex with X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
8B2F
 
 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | Descriptor: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
9DLF
 
 | | Arabinosyltransferase AftB in complex with Fab_B3 | | Descriptor: | Arabinosyltransferase AftB, Fab_B3 heavy chain, Fab_B3 light chain, ... | | Authors: | Liu, Y, Mancia, F. | | Deposit date: | 2024-09-10 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
8WT0
 
 | | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase | | Descriptor: | Cell filamentation protein Fic, D(-)-TARTARIC ACID, DUF2559 domain-containing protein, ... | | Authors: | Chen, F.R, Guo, L.W, Lu, C.H, Jiang, W.J, Luo, Z.Q, Liu, J.F, Zhang, L.Q. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase
To Be Published
|
|
6MZI
 
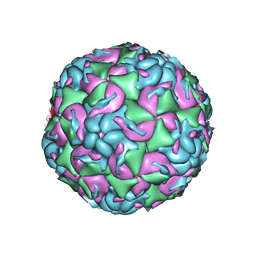 | | CryoEM structure of human enterovirus D68 expanded 1 particle (pH 6.5, 4 degrees Celsius, 3 min) | | Descriptor: | viral protein 1, viral protein 2, viral protein 3, ... | | Authors: | Liu, Y, Rossmann, M.G. | | Deposit date: | 2018-11-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for the acid-initiated uncoating of human enterovirus D68.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6Y1G
 
 | | Photoconverted HcRed in its optoacoustic state | | Descriptor: | 1,2-ETHANEDIOL, GFP-like non-fluorescent chromoprotein | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-02-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Challenging a Preconception: Optoacoustic Spectrum Differs from the Optical Absorption Spectrum of Proteins and Dyes for Molecular Imaging.
Anal.Chem., 92, 2020
|
|
6Y24
 
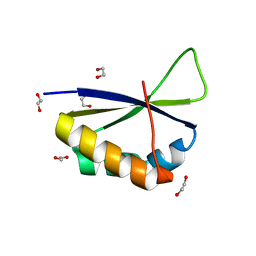 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6YI4
 
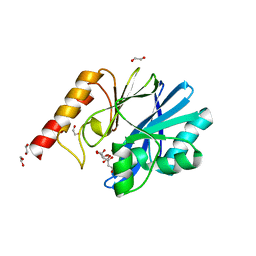 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
8GF1
 
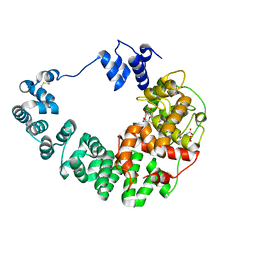 | |
7SCR
 
 | | Crystal structure of trypanosome brucei hypoxanthine-guanine-xanthine phosphoribzosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12068486 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
8GFC
 
 | |
7S4X
 
 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
6Y0Z
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6YPH
 
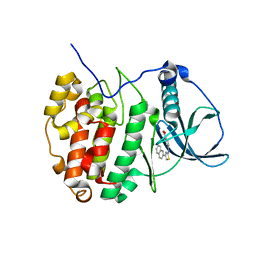 | | Crystal Structure of CK2alpha with Compound 2 bound | | Descriptor: | 4-[(4-naphthalen-2-yl-1,3-thiazol-2-yl)amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
8GFB
 
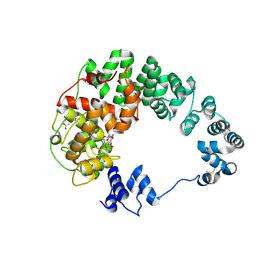 | |
6Q10
 
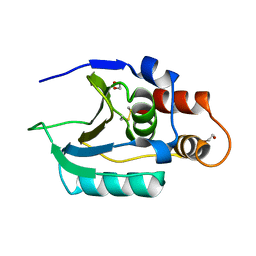 | |
434D
 
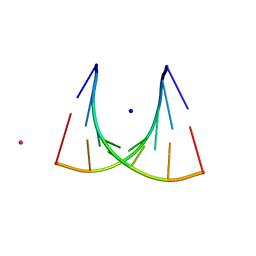 | | 5'-R(*UP*AP*GP*CP*UP*CP*C)-3', 5'-R(*GP*GP*GP*GP*CP*UP*A)-3' | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | Authors: | Mueller, U, Schuebel, H, Sprinzl, M, Heinemann, U. | | Deposit date: | 1998-10-23 | | Release date: | 1999-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution.
RNA, 5, 1999
|
|
6Q3P
 
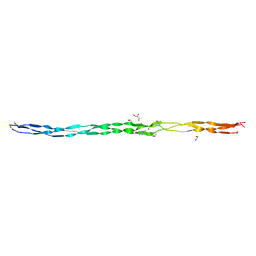 | | Atomic resolution crystal structure of an AAB collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading Chain of the AAB Collagen Heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
7PT2
 
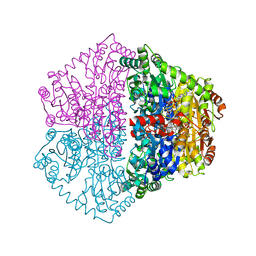 | |
6N64
 
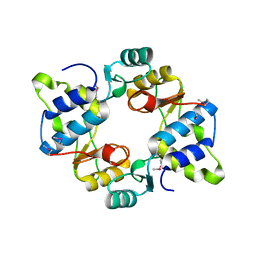 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
7PT1
 
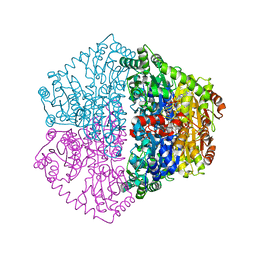 | |
8PH6
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(DPhF)(CO3)3] in condition B | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, Ru2(DPhF)(CO3)2(Formate), ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8DYS
 
 | | Crystal structure of human Eukaryotic translation initiation factor 2A (eIF2A) | | Descriptor: | 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 2A, GLYCEROL, ... | | Authors: | Righetto, G.L, Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Eukaryotic translation initiation factor 2A (eIF2A)
To Be Published
|
|
