8SFP
 
 | |
8SFO
 
 | |
8SFN
 
 | |
8SFM
 
 | | Crystal structure of the engineered SsoPox variant IVB10 in alternate state | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFL
 
 | |
8SFK
 
 | | Crystal structure of the engineered SsoPox variant IVE2 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFJ
 
 | | WT CRISPR-Cas12a with a 10bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*AP*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | WT CRISPR-Cas12a with a 10bp R-loop
To Be Published
|
|
8SFI
 
 | | WT CRISPR-Cas12a with a 8bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*TP*A)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | WT CRISPR-Cas12a with a 8bp R-loop
To Be Published
|
|
8SFH
 
 | |
8SFG
 
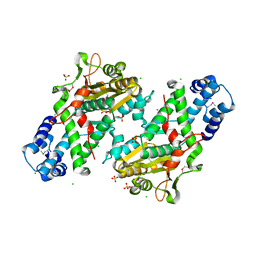 | | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6 | | Descriptor: | Autotransporter adhesin, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Herrera, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-04-11 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6.
To Be Published
|
|
8SFF
 
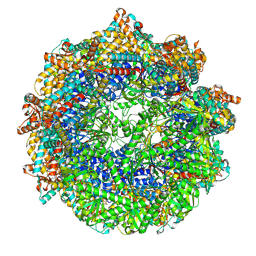 | | CCT G beta 5 complex closed state 0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SFE
 
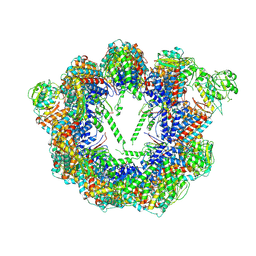 | | Open state CCT-G beta 5 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SFD
 
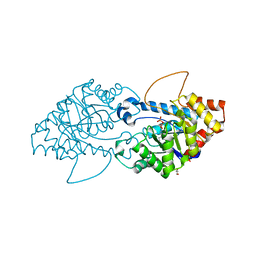 | | Crystal structure of the engineered SsoPox variant IVB10 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFC
 
 | | Crystal structure of the engineered SsoPox variant IVA4 in alternate state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFB
 
 | | Crystal structure of the engineered SsoPox variant IVA4 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFA
 
 | | Crystal structure of the engineered SsoPox variant IIIC1 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF9
 
 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF8
 
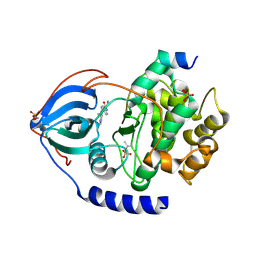 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
8SF7
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain MEC17 | | Descriptor: | CFAM166A, CFAM166B, CFAM166C, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effect of alpha-tubulin acetylation on the doublet microtubule structure.
Elife, 12, 2024
|
|
8SF6
 
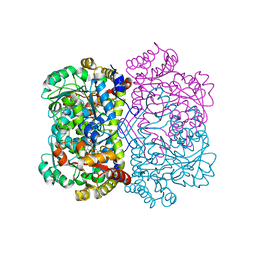 | | Promiscuous amino acid gamma synthase from Caldicellulosiruptor hydrothermalis in closed conformation | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Buller, A.R, Zmich, A.P, Bingman, C.A. | | Deposit date: | 2023-04-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Multiplexed Assessment of Promiscuous Non-Canonical Amino Acid Synthase Activity in a Pyridoxal Phosphate-Dependent Protein Family.
Acs Catalysis, 13, 2023
|
|
8SF5
 
 | |
8SF4
 
 | |
8SF3
 
 | |
8SF2
 
 | | Crystal structure of the engineered SsoPox variant IG7 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF1
 
 | | Carbonic anhydrase II XFEL radiation damage RT | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
