5N7G
 
 | | MAGI-1 complexed with a synthetic pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
5X6V
 
 | | Crystal structure of human heteroheptameric complex | | Descriptor: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
5XM2
 
 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
8HXZ
 
 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HXX
 
 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PFX
 
 | |
7PEX
 
 | |
7PFC
 
 | |
7PEZ
 
 | |
7PF3
 
 | |
7PF5
 
 | |
7PET
 
 | | The 4x177 nucleosome array containing H1 | | Descriptor: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | Authors: | Dombrowski, M, Cramer, P. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PF6
 
 | |
7PFU
 
 | |
7PF2
 
 | |
7PFA
 
 | |
7PEU
 
 | |
7PFD
 
 | |
7PFE
 
 | |
7PFT
 
 | |
7PF0
 
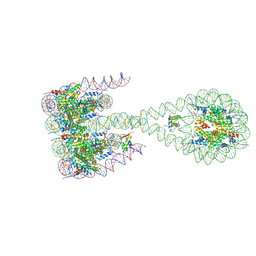 | |
7PFV
 
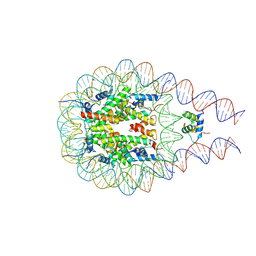 | |
7PFW
 
 | |
6G0L
 
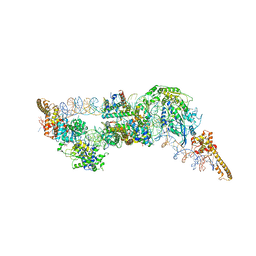 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
1R0E
 
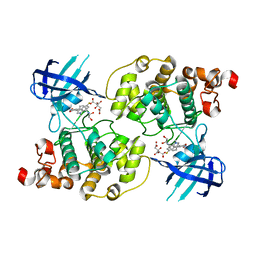 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
