6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
7Q4F
 
 | | Structure of coproheme decarboxylase from Corynebacterium dipththeriae W183Y mutant in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Coproheme decarboxylase from Corynebacterium dipththeriae W183Y mutant in complex with coproheme, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michlits, H, Valente, N, Mlynek, G, Hofbauer, S. | | Deposit date: | 2021-10-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Initial Steps to Engineer Coproheme Decarboxylase to Obtain Stereospecific Monovinyl, Monopropionyl Deuterohemes.
Front Bioeng Biotechnol, 9, 2021
|
|
4WZL
 
 | |
4WZV
 
 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
4X0L
 
 | | Human haptoglobin-haemoglobin complex | | Descriptor: | CACODYLATE ION, GLYCEROL, Haptoglobin, ... | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4WMB
 
 | |
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
6W4V
 
 | | Structure of anti-ferroportin Fab45D8 | | Descriptor: | 1,2-ETHANEDIOL, Fab45D8 Heavy Chain, Fab45D8 Light Chain | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
4X1X
 
 | |
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
6VXI
 
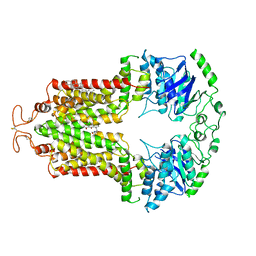 | | Structure of ABCG2 bound to mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
6WAX
 
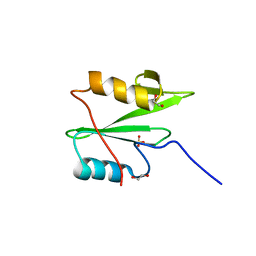 | | C-terminal SH2 domain of p120RasGAP | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6YPI
 
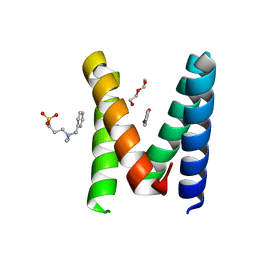 | | Structure of the engineered metallo-Diels-Alderase DA7 W16G,K58Q,L77R,T78R | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BENZOIC ACID, DA7 W16G,K58Q,L77R,T78R, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
7S85
 
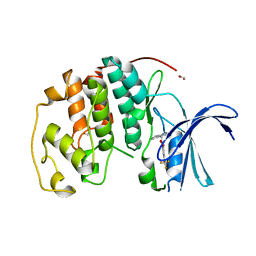 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
6W11
 
 | |
4X3C
 
 | |
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
7CV3
 
 | | Quadruplex-duplex hybrid structure in the PIM1 gene, Form 1 | | Descriptor: | PIM1 promoter, Form 1 | | Authors: | Winnerdy, F.R, Tan, D.J.Y, Lim, K.W, Phan, A.T. | | Deposit date: | 2020-08-25 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two quadruplex-duplex hybrids in the PIM1 gene.
Nucleic Acids Res., 48, 2020
|
|
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
8J43
 
 | | Reductive aminase RA29-WT | | Descriptor: | 1,2-ETHANEDIOL, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
6WKD
 
 | | Crystal structure of pentalenene synthase complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
8RI3
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 7 days of soaking | | Descriptor: | AMMONIA, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
6WHI
 
 | | Cryo-electron microscopy structure of the type I-F CRISPR RNA-guided surveillance complex bound to the anti-CRISPR AcrIF9 | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-04-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
