5WED
 
 | |
5WFK
 
 | |
5WLJ
 
 | |
5WF0
 
 | |
5WLM
 
 | |
5WMB
 
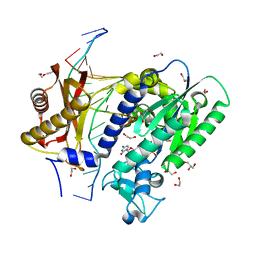 | | Structure of the 10S (-)-cis-BP-dG modified Rev1 ternary complex (the BP residue is disordered) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
5WMD
 
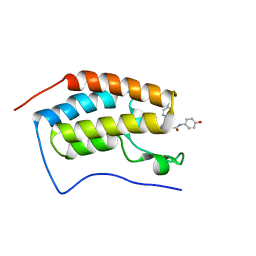 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-hydroxyphenyl)acetamide, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5M03
 
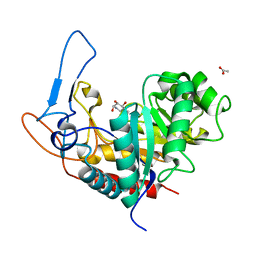 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5WMV
 
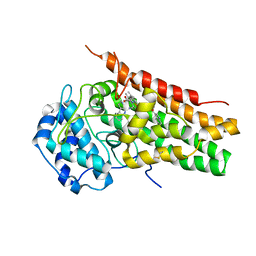 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WMM
 
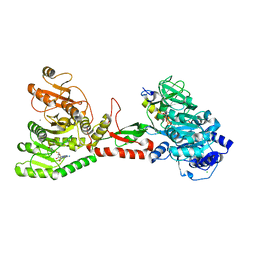 | | Crystal structure of an adenylation domain interrupted by a methylation domain (AMA4) from nonribosomal peptide synthetase TioS | | Descriptor: | (2S)-2-amino-3-methylbutanoyl (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl hydrogen (S)-phosphate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pang, A.H, Mori, S, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-07-30 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for backbone N-methylation by an interrupted adenylation domain.
Nat. Chem. Biol., 14, 2018
|
|
5WNZ
 
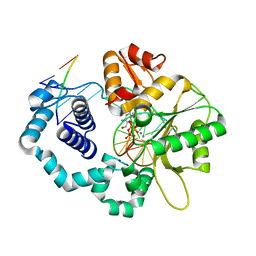 | | DNA polymerase beta substrate complex with incoming 5-FodCTP | | Descriptor: | 2'-deoxy-5-formylcytidine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5WO0
 
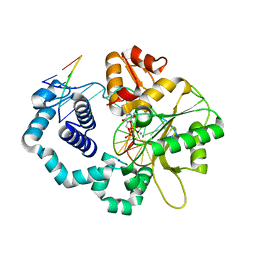 | | DNA polymerase beta substrate complex with incoming 5-FodUTP | | Descriptor: | 2'-deoxy-5-formyluridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
7R5I
 
 | | High resolution Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein | | Descriptor: | CALCIUM ION, Exosporium protein, GLN-GLU-ASP-PHE-SER-SER-ASP-SER-SER-PHE-SER, ... | | Authors: | Brear, P, Schack, S, Christie, G. | | Deposit date: | 2022-02-10 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein"
To Be Published
|
|
5WO8
 
 | |
5WQ0
 
 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
5WTF
 
 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LHD
 
 | | Structure of glycosylated human aminopeptidase N | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Mudgal, G, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2016-07-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5WQN
 
 | |
5WQX
 
 | | Covalent bond formation of synthetic ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-2-oxidanylidenehexadec-5-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
5WRM
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WSF
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5WWF
 
 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WSP
 
 | | Crystal structure of DNA3 duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), STRONTIUM ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSR
 
 | | Crystal structure of T-Hg-T pair containing DNA duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
