9BH6
 
 | |
5N02
 
 | | Crystal structure of the decarboxylase AibA/AibB C56S variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2JST
 
 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
9BHA
 
 | |
5NJR
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 50s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
7RP8
 
 | |
1AG0
 
 | |
9BHR
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-{(1E)-3-[(2-methoxyethyl)amino]-3-oxoprop-1-en-1-yl}-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1 | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Krausser, C, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound
To be published
|
|
5BYW
 
 | |
6RKI
 
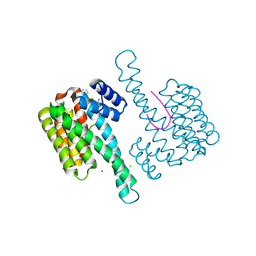 | | Fragment AZ-023 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[(3-aminophenyl)amino]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
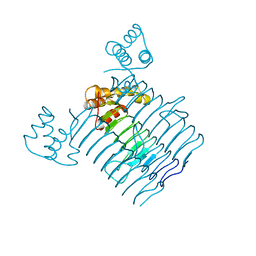
3HSQ
 
 | |
6RKM
 
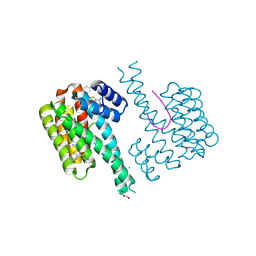 | | Fragment AZ-022 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-phenylazanyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5N5M
 
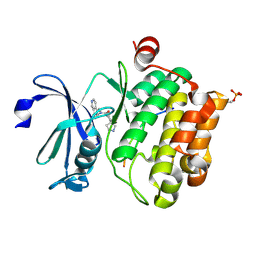 | |
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
5N5W
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV3 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyaniline, CALCIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5NLD
 
 | | Chicken GRIFIN (crystallisation pH: 7.5) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
6RL4
 
 | | Fragment AZ-025 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[1-(2-azanylethyl)imidazol-4-yl]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
3P6P
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio | | Descriptor: | (3S,5S,7S)-N-[6-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexyl]tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio
To be Published
|
|
2C2U
 
 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, SULFATE ION, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-30 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
3P04
 
 | | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8 | | Descriptor: | Uncharacterized BCR | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Hamilton, H, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8.
To be Published
|
|
5CHG
 
 | |
9EHI
 
 | |
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
7S4F
 
 | | Protein Tyrosine Phosphatase 1B - F182Q mutant bound with Hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brandao, T.A.S, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
1EB8
 
 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-ACETONE-CYANOHYDRIN LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
