7MAI
 
 | | HIV-1 Protease (I84V) in Complex with PU5 (LR4-47) | | Descriptor: | Protease, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl][(2S)-2-methylbutyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU5 (LR4-47)
To Be Published
|
|
8VBF
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (40-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
7M9R
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR4-44 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butan-2-yl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with LR4-44
To Be Published
|
|
6W8S
 
 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
8VBI
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBE
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (39-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
7M9O
 
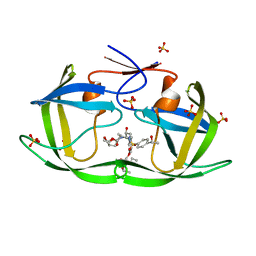 | | HIV-1 Protease (I84V) in Complex with LR3-48 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1R)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9Q
 
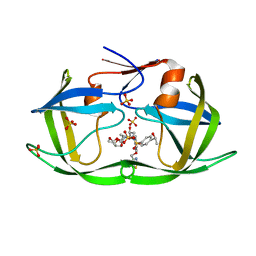 | | HIV-1 Protease WT (NL4-3) in Complex with LR4-33 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]-1-{4-[(2-methyl-1,3-thiazol-4-yl)methoxy]phenyl}butan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with LR4-33
To Be Published
|
|
6W2O
 
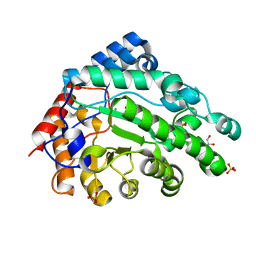 | |
8VC9
 
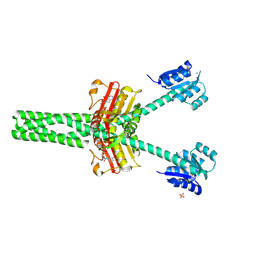 | |
8VBH
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
7MAL
 
 | | HIV-1 Protease (I84V) in Complex with PU8 (LR4-06) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU8 (LR4-06)
To Be Published
|
|
5I1D
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
6W9O
 
 | |
7MAO
 
 | | HIV-1 Protease (I84V) in Complex with PU10 (LR4-07) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU10 (LR4-07)
To Be Published
|
|
8VJZ
 
 | | HLA-A*03:01 with WT KRAS-10mer | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GTPase KRas, ... | | Authors: | Sim, M.J.W, Sun, P.D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of a mutant KRAS-G12V specific TCR restricted by HLA-A3.
Eur.J.Immunol., 54, 2024
|
|
7M9F
 
 | |
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
6W9R
 
 | |
8VZG
 
 | | Carbonic anhydrase II mutant with DRD function | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CITRATE ANION, Carbonic anhydrase 2, ... | | Authors: | Sethi, D.K. | | Deposit date: | 2024-02-11 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Functional regulation of membrane-bound cytokine activity in engineered T cells via drug-responsive domains derived from carbonic anhydrase 2
To Be Published
|
|
8VFM
 
 | | Salmonella effector protein SipA decorated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Guo, E.Z, Galan, J.E. | | Deposit date: | 2023-12-21 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacterial effector protein SipA bound to F-actin reveals a unique mechanism for filament stabilization.
Biorxiv, 2024
|
|
5VHW
 
 | | GluA2-0xGSG1L bound to ZK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
7M9M
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-55 | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
5I7Y
 
 | | BRD9 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4514 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
6W3E
 
 | | Structure of phosphorylated IRE1 in complex with G-0701 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, methyl ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]carbamate | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.737 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
