4PET
 
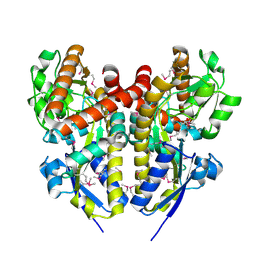 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM COLWELLIA PSYCHRERYTHRAEA (CPS_0129, TARGET EFI-510097) WITH BOUND CALCIUM AND PYRUVATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PLZ
 
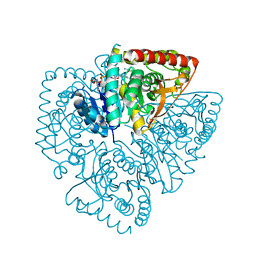 | | Crystal structure of Plasmodium falciparum lactate dehydrogenase mutant W107fA. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase, OXAMIC ACID | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PM7
 
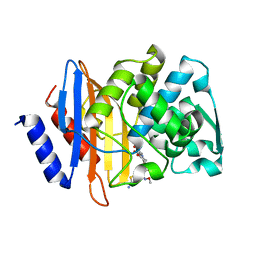 | | Crystal structure of CTX-M-14 S70G:S237A in complex with cefotaxime at 1.29 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4LNH
 
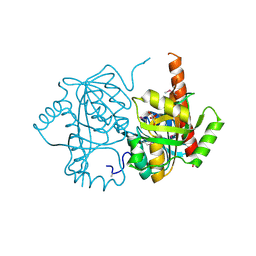 | | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520. | | Descriptor: | GLYCEROL, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520.
To be Published
|
|
1TAB
 
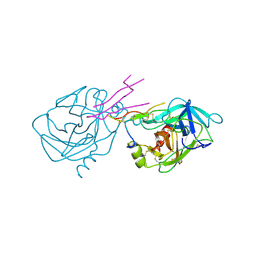 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
3W8O
 
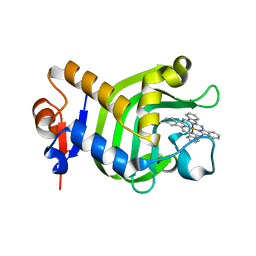 | | Crystal Structure of HasAp with Iron Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, [29H,31H-phthalocyaninato(2-)-kappa~4~N~29~,N~30~,N~31~,N~32~]iron | | Authors: | Shirataki, C, Shoji, O, Terada, M, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-20 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3ZJ7
 
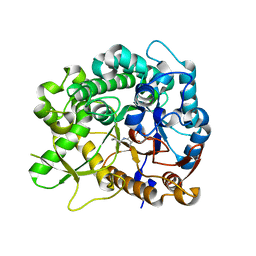 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
4LBG
 
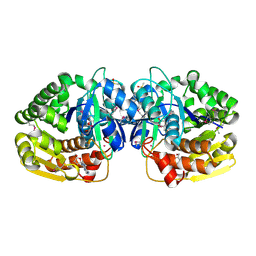 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine
To be Published
|
|
3ZCX
 
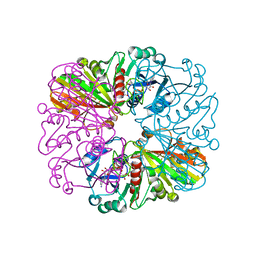 | |
5RHG
 
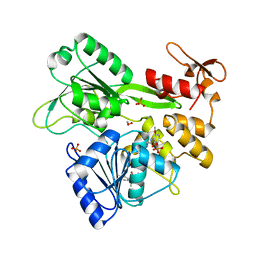 | |
4PPV
 
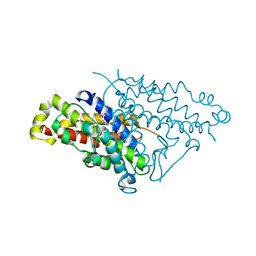 | |
5RHW
 
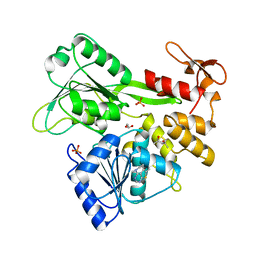 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z31222641 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NS3 Helicase, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3ZBJ
 
 | | Fitting results in the I-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
4PKQ
 
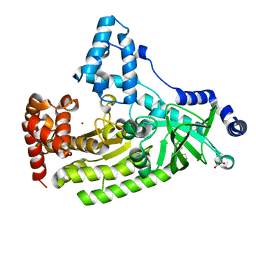 | | Anthrax toxin lethal factor with bound zinc | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, TRIETHYLENE GLYCOL, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1Y6Z
 
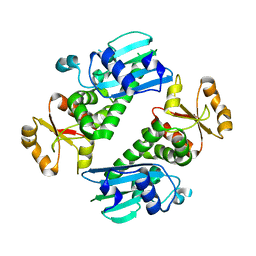 | | Middle domain of Plasmodium falciparum putative heat shock protein PF14_0417 | | Descriptor: | heat shock protein, putative | | Authors: | Lunin, V.V, Botchkareva, E, Loppnau, P, Amani, M, Bray, J, Vedadi, M, Edwards, A, Arrowsmith, C, Sundstrom, M, Bochkarev, A, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-07 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3ZCG
 
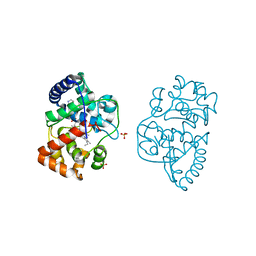 | | Ascorbate peroxidase W41A-H42C mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of a Heme Peroxidase.
Dalton Trans, 42, 2013
|
|
4PM9
 
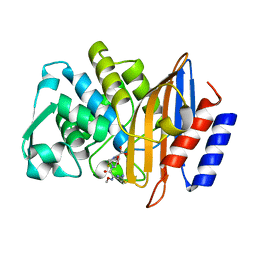 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase in complex with cefotaxime at 1.45 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Cardenas, A.M, Adamski, C.J, Sankaran, B, Brown, N.G, Horton, L.B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PMJ
 
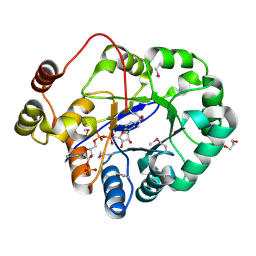 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
1UXX
 
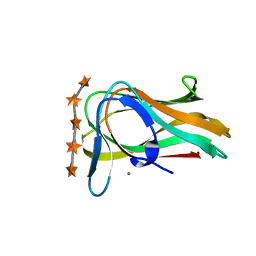 | | CBM6ct from Clostridium thermocellum in complex with xylopentaose | | Descriptor: | CALCIUM ION, XYLANASE U, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Henrissat, D.B.B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1YCM
 
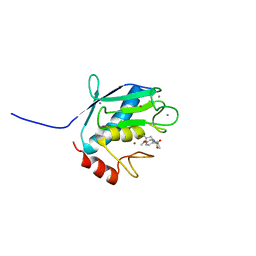 | | Solution Structure of matrix metalloproteinase 12 (MMP12) in the presence of N-Isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UYK
 
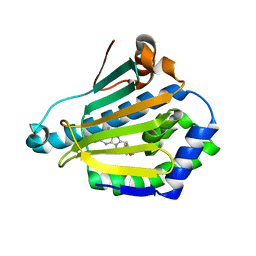 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
4IZH
 
 | |
4PFH
 
 | | Crystal structure of engineered D-tagatose 3-epimerase PcDTE-IDF8 | | Descriptor: | D-fructose, D-psicose, D-tagatose 3-epimerase, ... | | Authors: | Hee, C.S, Bosshart, A, Schirmer, T. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Divergent Evolution of a Thermostable D-Tagatose Epimerase towards Improved Activity for Two Hexose Substrates.
Chembiochem, 16, 2015
|
|
4PON
 
 | | The crystal structure of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis | | Descriptor: | Putative RNA methylase | | Authors: | Park, S.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4PQH
 
 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
