5AKD
 
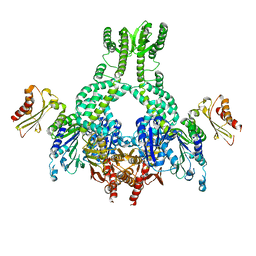 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
1IDL
 
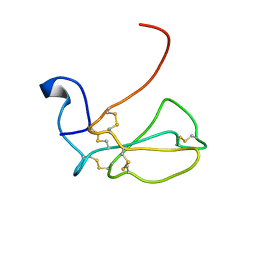 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDG
 
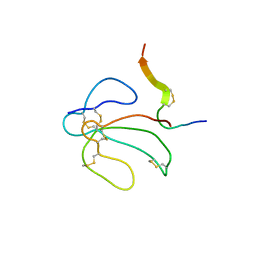 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1W8G
 
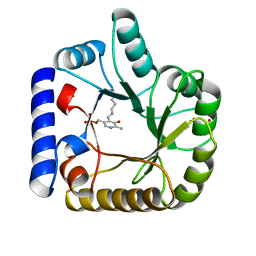 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
1GV3
 
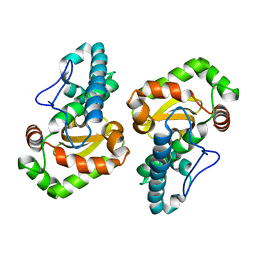 | | The 2.0 Angstrom resolution structure of the catalytic portion of a cyanobacterial membrane-bound manganese superoxide dismutase | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Atzenhofer, W, Regelsberger, G, Jacob, U, Huber, R, Peschek, G.A, Obinger, C. | | Deposit date: | 2002-02-05 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Structure of the Catalytic Portion of a Cyanobacterial Membrane-Bound Manganese Superoxide Dismutase
J.Mol.Biol., 321, 2002
|
|
2RO1
 
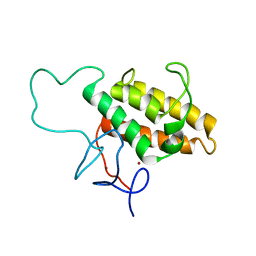 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
1UXT
 
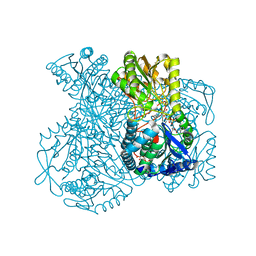 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXN
 
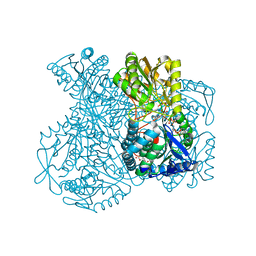 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1IDI
 
 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1HM8
 
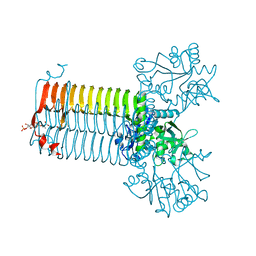 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1UXU
 
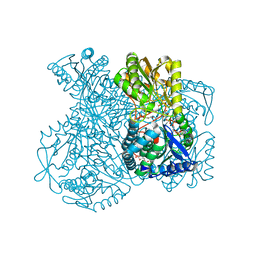 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1HM0
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE, GLMU | | Descriptor: | CALCIUM ION, N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1HL8
 
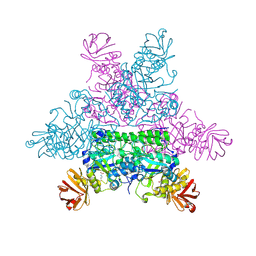 | |
1D2N
 
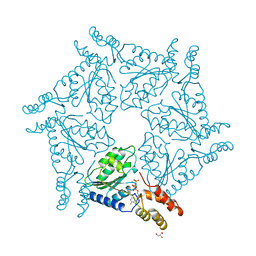 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1UXP
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1XBP
 
 | | Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with Tiamulin | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Schluenzen, F, Pyetan, E, Fucini, P, Yonath, A, Harms, J.M. | | Deposit date: | 2004-08-31 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin.
Mol.Microbiol., 54, 2004
|
|
1UXQ
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1HL9
 
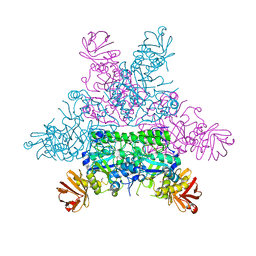 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH A MECHANISM BASED INHIBITOR | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, PUTATIVE ALPHA-L-FUCOSIDASE | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1HM9
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A AND UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, ... | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
2RNW
 
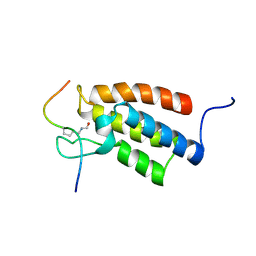 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
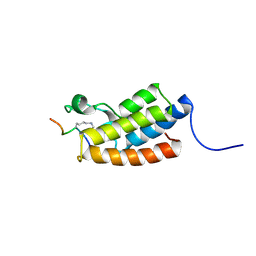 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNY
 
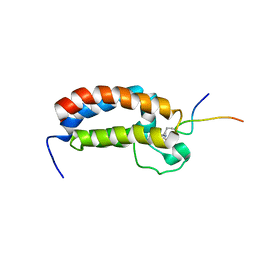 | | Complex Structures of CBP Bromodomain with H4 ack20 Peptide | | Descriptor: | CREB-binding protein, Histone H4 | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
1UXR
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
