6XPE
 
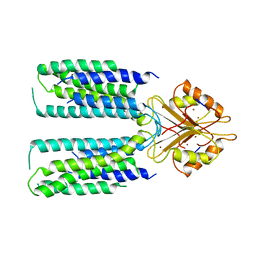 | | Cryo-EM structure of human ZnT8 WT, in the presence of zinc, determined in outward-facing conformation | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Bai, X.C, Xue, J, Jiang, Y.X. | | Deposit date: | 2020-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations.
Elife, 9, 2020
|
|
6XUE
 
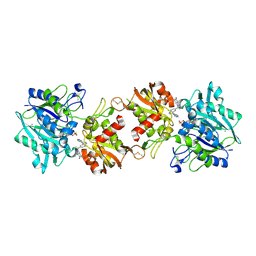 | |
6XYC
 
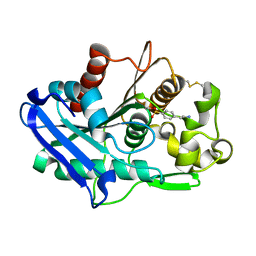 | | Truncated form of carbohydrate esterase from gut microbiota | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Acetyl xylan esterase | | Authors: | Penttinen, L, Hakulinen, N, Master, E. | | Deposit date: | 2020-01-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
6XPD
 
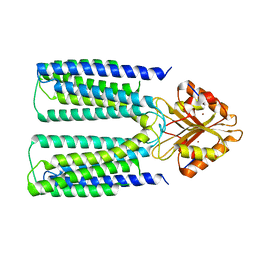 | |
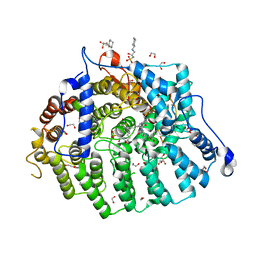
8T70
 
 | |
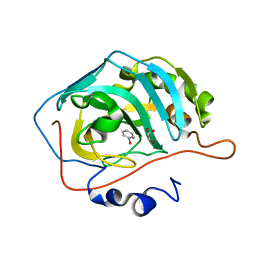
6XWZ
 
 | |
8SGT
 
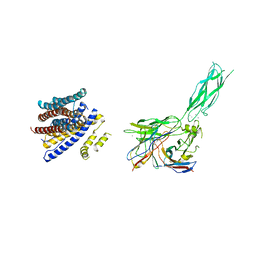 | |
8SZV
 
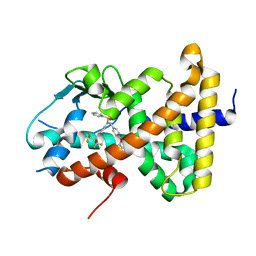 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
8SGJ
 
 | |
2V9C
 
 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
8SJ5
 
 | | Walnut Tree Phytocystatin | | Descriptor: | Cysteine proteinase inhibitor | | Authors: | Valadares, N.F. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and interconversion of monomers and domain-swapped dimers of the walnut tree phytocystatin.
Biochim Biophys Acta Proteins Proteom, 1872, 2023
|
|
6XKQ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6XPF
 
 | | Cryo-EM structure of human ZnT8 WT, in the absence of zinc, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Bai, X.C, Xue, J, Jiang, Y.X. | | Deposit date: | 2020-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations.
Elife, 9, 2020
|
|
1AZ8
 
 | | BOVINE TRYPSIN COMPLEXED TO BIS-PHENYLAMIDINE INHIBITOR | | Descriptor: | +/-METHYL 4-(AMINOIMINOMETHYL)-BETA-[3- INH (AMINOIMINO)PHENYL]BENZENE PENTANOATE, TRYPSIN | | Authors: | Alexander, R, Smallwood, A. | | Deposit date: | 1997-11-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unexpected Binding Mode of a Bis-Phenylamidine Factor Xa Inhibitor Complexed to Bovine Trypsin
To be Published
|
|
8RDZ
 
 | |
6YBK
 
 | | Structure of MBP-Mcl-1 in complex with compound 4d | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-(pyrazin-2-ylmethoxy)phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
8SGI
 
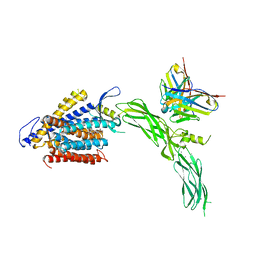 | | Cryo-EM structure of human NCX1 in complex with SEA0400 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, CALCIUM ION, Fab heavy chain, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms of human cardiac sodium calcium exchanger NCX1
To Be Published
|
|
2Z98
 
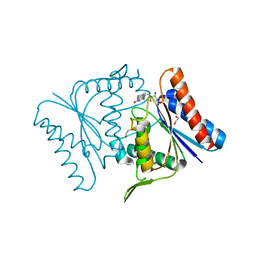 | |
2Z9C
 
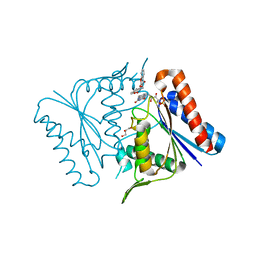 | |
6YCQ
 
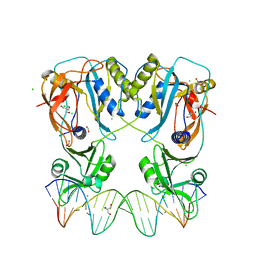 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | Authors: | Crespo, I, Weijers, D, Boer, D.R. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2Z9D
 
 | |
8BGI
 
 | | GABA-A receptor a5 homomer - a5V1 - Flumazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, Pregnanolone, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BOE
 
 | |
2A32
 
 | | Trypsin in complex with benzene boronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, BORATE ION, CALCIUM ION, ... | | Authors: | Transue, T.R, Gabel, S.A, London, R.E. | | Deposit date: | 2005-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NMR and crystallographic characterization of adventitious borate binding by trypsin.
Bioconjug.Chem., 17, 2006
|
|
