8OE5
 
 | |
6AQU
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AVC
 
 | |
8OE1
 
 | |
7UR6
 
 | |
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8DI7
 
 | | CMY-2 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ahmadvand, P, Call, D.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural characterization of CMY-2 and its interactions with ampicillin and the cephalosporins - ceftiofur, DFC, DFC-dimer, DFC-cysteine, and nitrocefin
To Be Published
|
|
8P5V
 
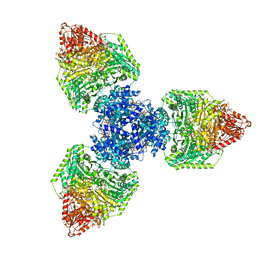 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
6ATU
 
 | |
6AS7
 
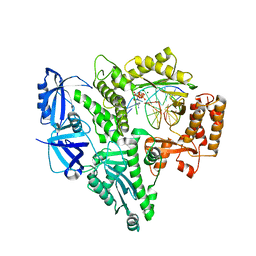 | | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF HUMAN DNA POLYMERASE ALPHA IN TERNARY COMPLEX WITH AN DNA-PRIMED DNA TEMPLATE AND DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, COBALT (II) ION, DNA (5'-D(*AP*GP*GP*CP*GP*CP*TP*CP*CP*AP*GP*GP*C)-3'), ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Activity and fidelity of human DNA polymerase alpha depend on primer structure.
J. Biol. Chem., 293, 2018
|
|
8P67
 
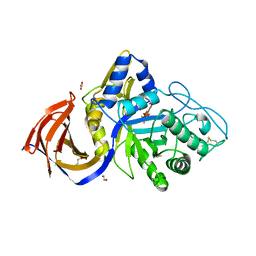 | | Crystal structure of Thermothelomyces thermophila (double mutant EE) in complex with aldotetrauronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GH30 family xylanase, ... | | Authors: | Dimarogona, M, Pentari, C, Kosinas, C, Topakas, E. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and molecular insights into a bifunctional glycoside hydrolase 30 xylanase specific to glucuronoxylan.
Biotechnol.Bioeng., 121, 2024
|
|
7SZ2
 
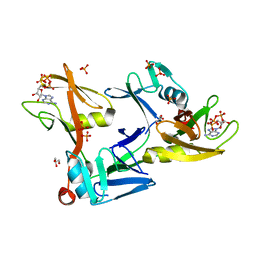 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
8DOZ
 
 | |
8P7G
 
 | |
8PWU
 
 | |
5XTM
 
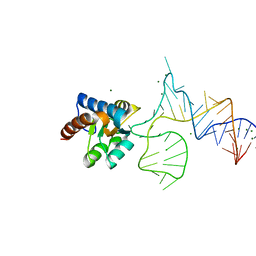 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.2 helix | | Descriptor: | 50S ribosomal protein L7Ae, MAGNESIUM ION, RNA (47-MER) | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-06-20 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5XO3
 
 | |
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
7SZ3
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ADPr | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
5XQP
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7UJ3
 
 | |
7UVG
 
 | |
7UOJ
 
 | |
8E2C
 
 | |
7UH9
 
 | | NMR structure of the cNTnC-cTnI chimera bound to W8 | | Descriptor: | CALCIUM ION, N-(7-aminoheptyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
